2022

Hoffman, Shannon M., Makoto A. Lalwani, and Jose L. Avalos. “Light-Controlled Fermentations for Microbial Chemical and Protein Production”. Journal of Visualized Experiments (2022). Web.
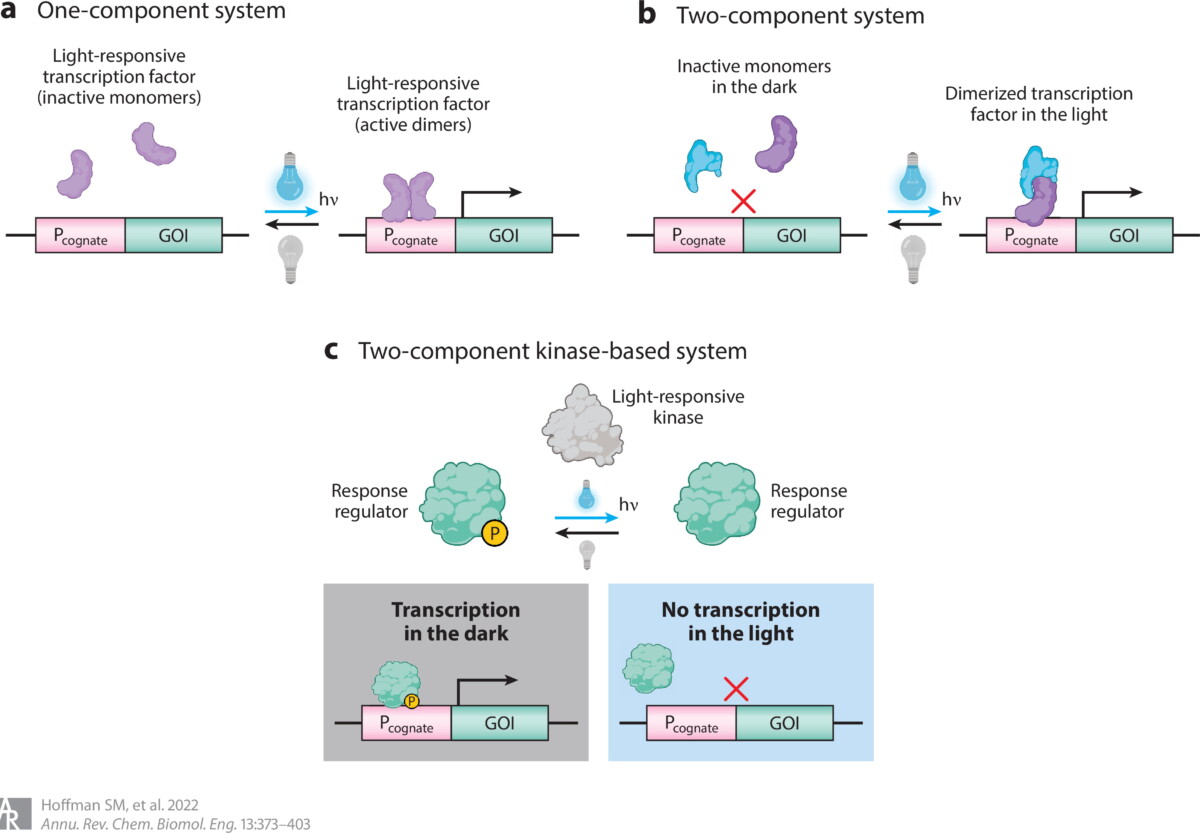
Hoffman, Shannon M., Allison Y. Tang, and José L. Avalos. “Optogenetics Illuminates Applications in Microbial Engineering.” Annual Review of Chemical and Biomolecular Engineering 13 (2022).
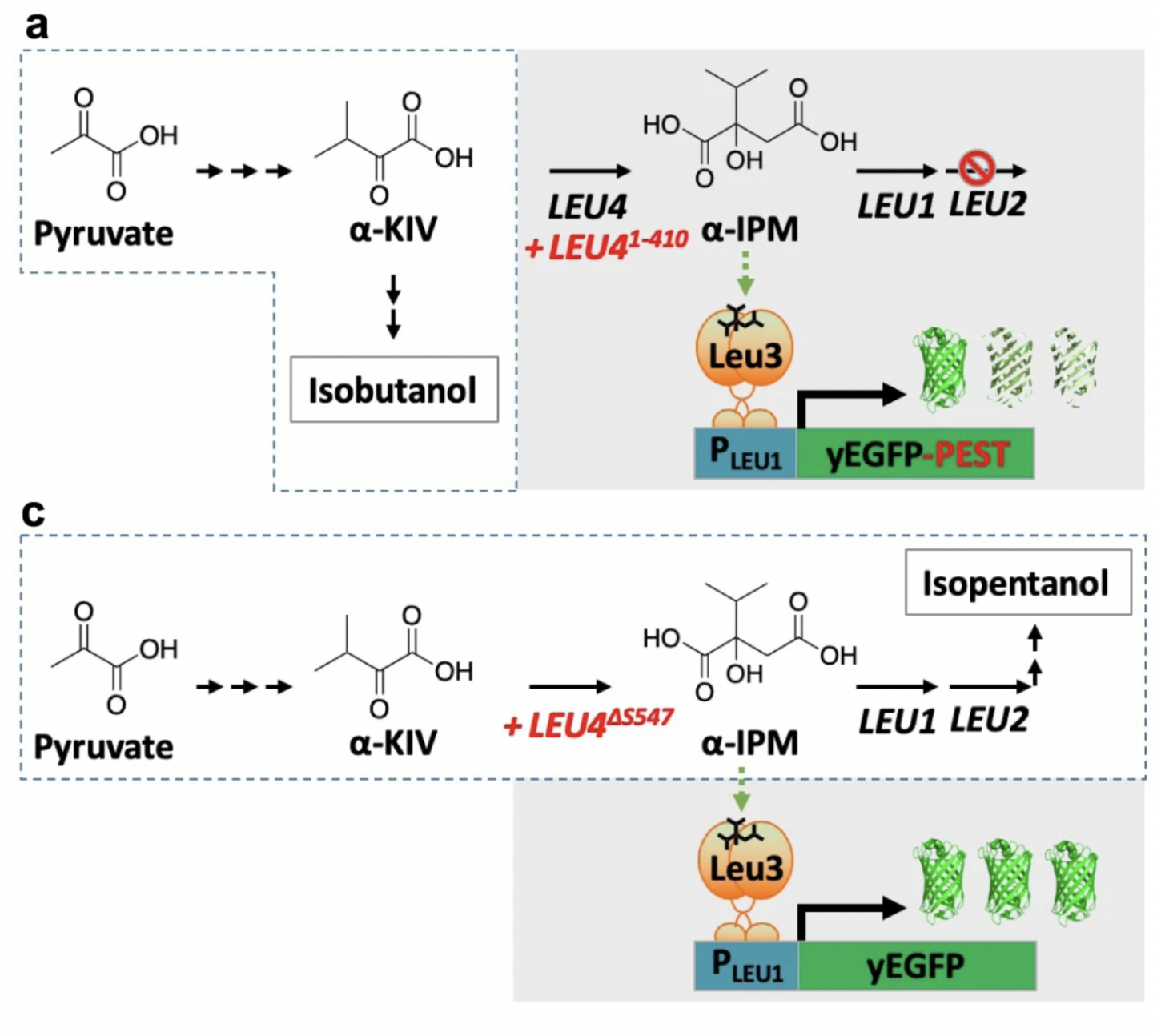
Zhang, Yanfei, et al. “Biosensor for branched-chain amino acid metabolism in yeast and applications in isobutanol and isopentanol production”. Nature Communications (2022). Web.
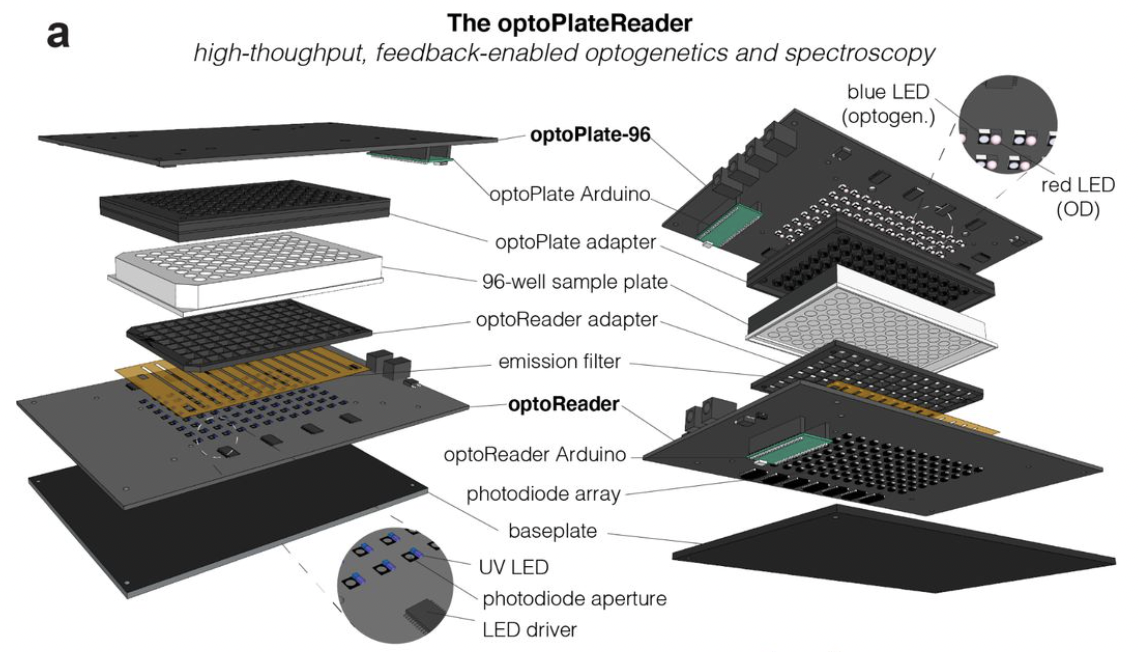
Datta, Saachi, William Benman, David Gonzalez-Martinez, Gloria Lee, Juliette Hooper, Grace Qian, Gabrielle Leavitt et al. “High-throughput feedback-enabled optogenetic stimulation and spectroscopy in microwell plates.” bioRxiv (2022).
López, José Montaño, Lisset Duran, and José L. Avalos. “Physiological limitations and opportunities in microbial metabolic engineering”. Nature Reviews Microbiology (2022). Web.
2021
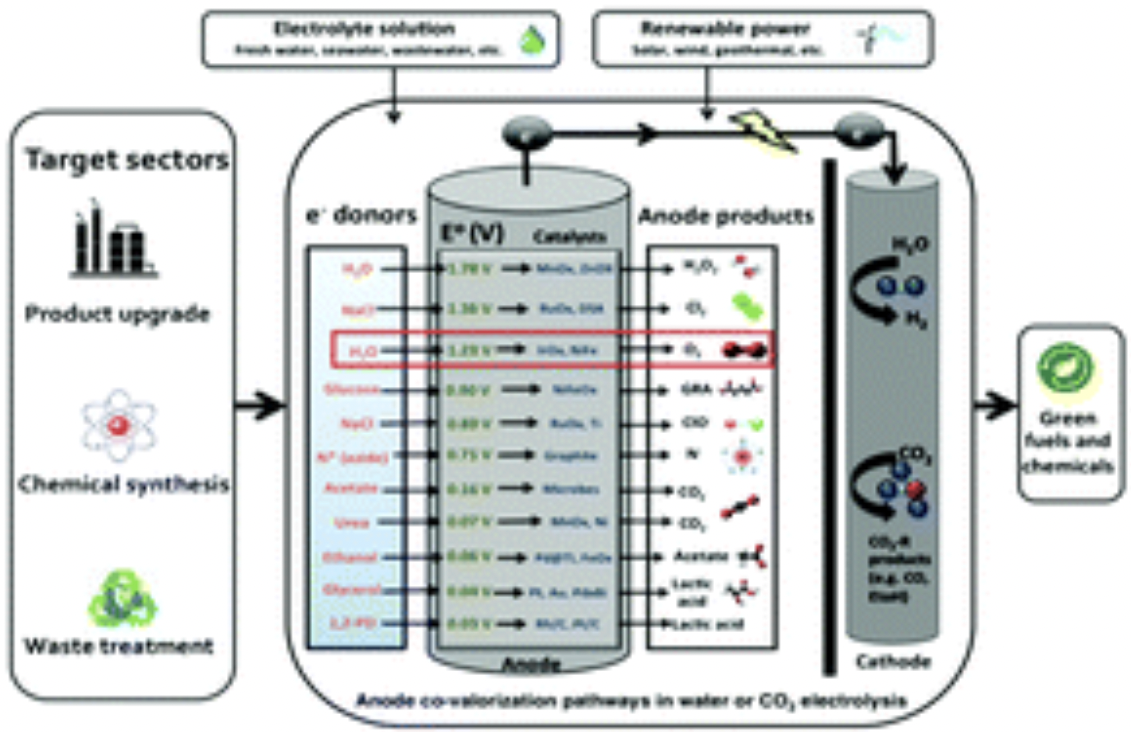
Jack, Joshua, et al. “Anode co-valorization for scalable and sustainable electrolysis”. Green Chemistry (2021). Web.
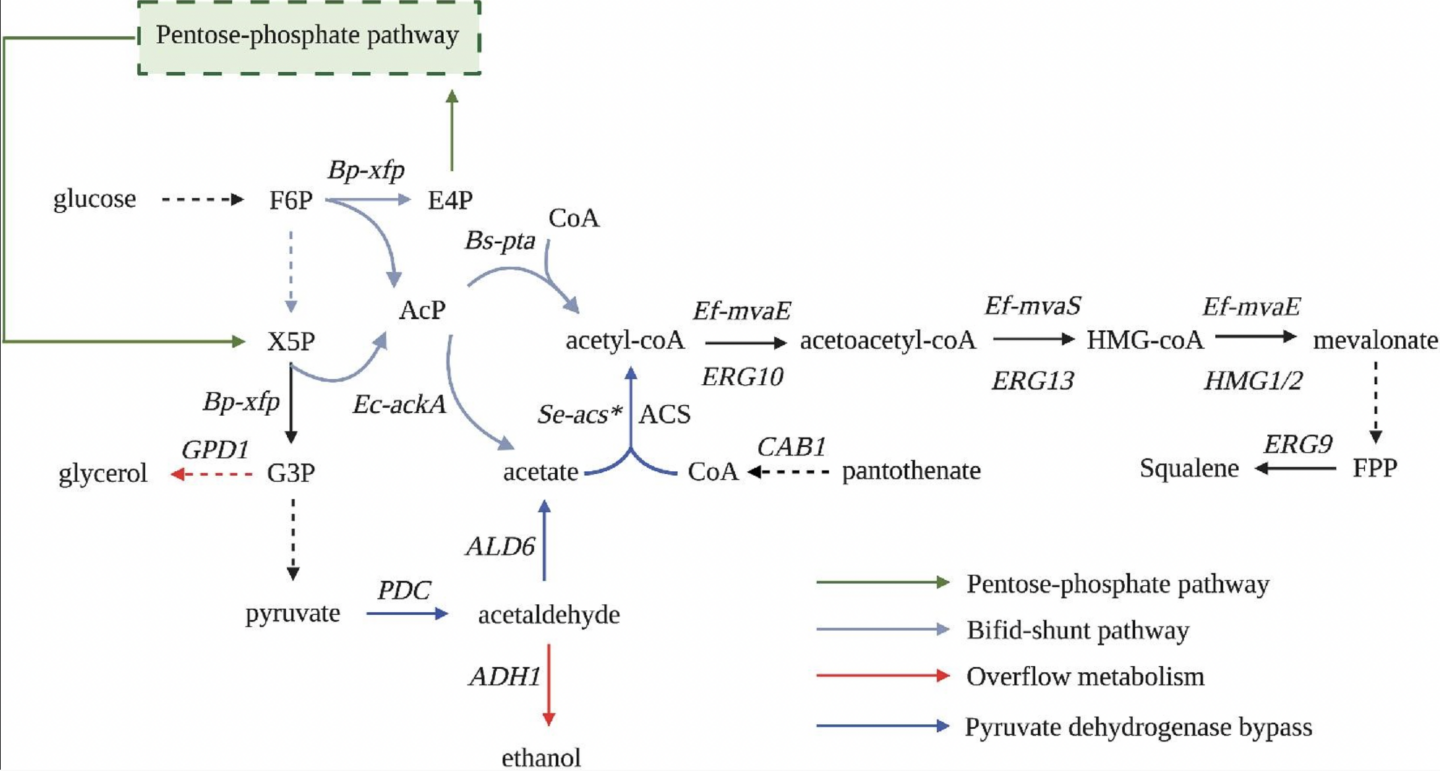
Wegner, Scott A., et al. “Engineering acetyl-CoA supply and ERG9 repression to enhance mevalonate production in Saccharomyces cerevisiae”. Journal of Industrial Microbiology and Biotechnology (2021). Print.
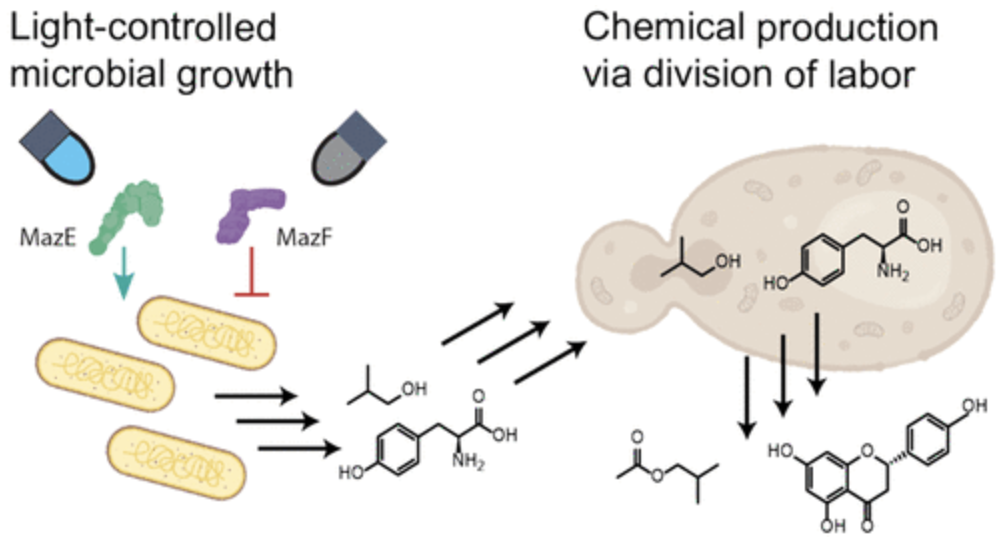
Lalwani, Makoto A., et al. “Optogenetic Control of Microbial Consortia Populations for Chemical Production”. ACS Synthetic Biology (2021). Web.
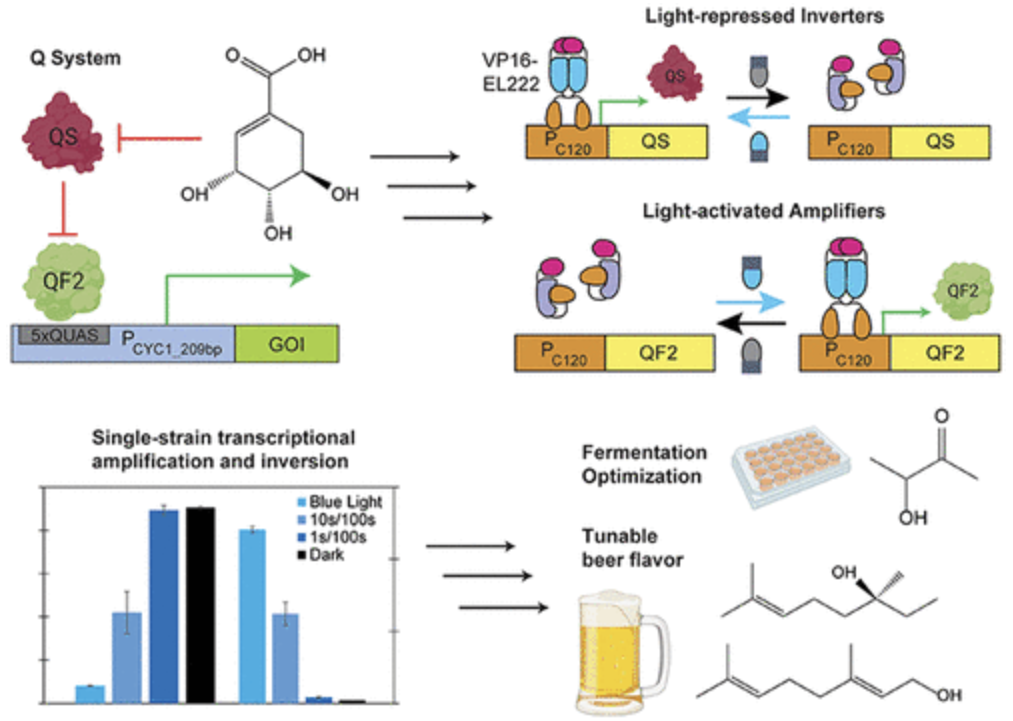
Lalwani, Makoto A., et al. “The Neurospora crassa Inducible Q System Enables Simultaneous Optogenetic Amplification and Inversion in Saccharomyces cerevisiae for Bidirectional Control of Gene Expression”. ACS Synthetic Biology (2021). Web.
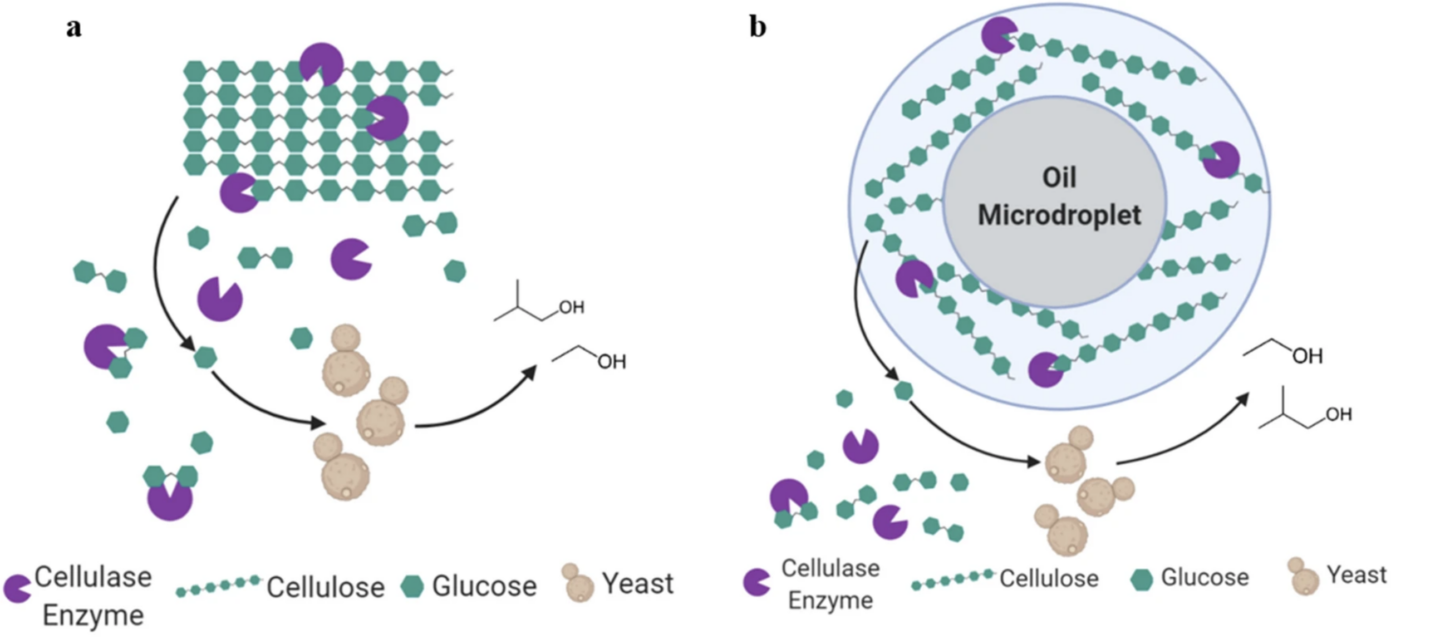
Hoffman, Shannon M., et al. “Cellulosic biofuel production using emulsified simultaneous saccharification and fermentation (eSSF) with conventional and thermotolerant yeasts”. Biotechnology for Biofuels (2021). Web.
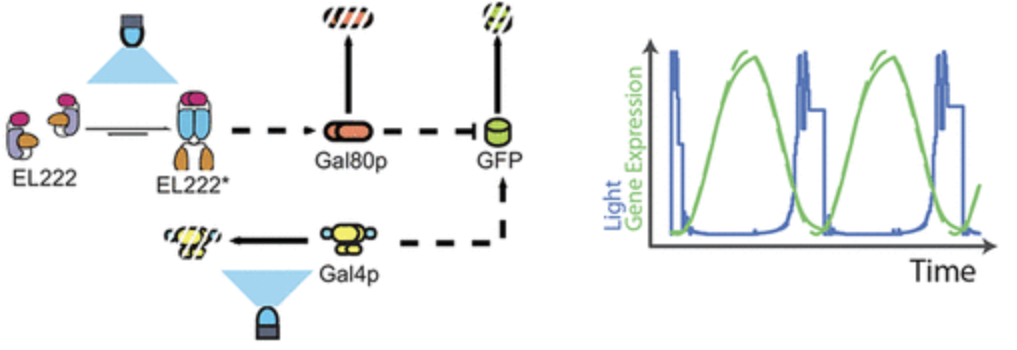
Lovelett, Robert J., et al. “Dynamical Modeling of Optogenetic Circuits in Yeast for Metabolic Engineering Applications”. ACS Synthetic Biology 10.2 (2021): , 10, 2, 219–227. Web.
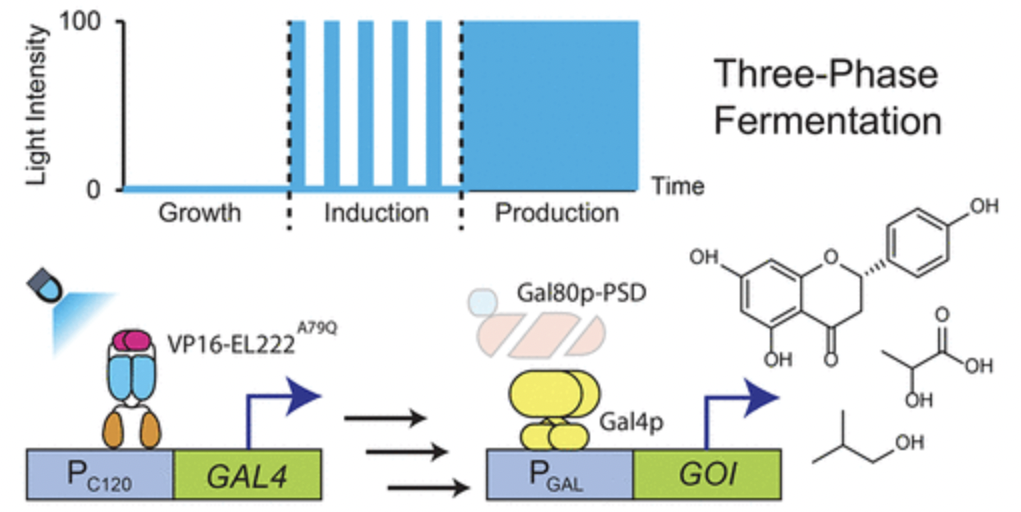
Zhao, Evan M., et al. “Optogenetic Amplification Circuits for Light-Induced Metabolic Control”. ACS Synthetic Biology (2021). Web.
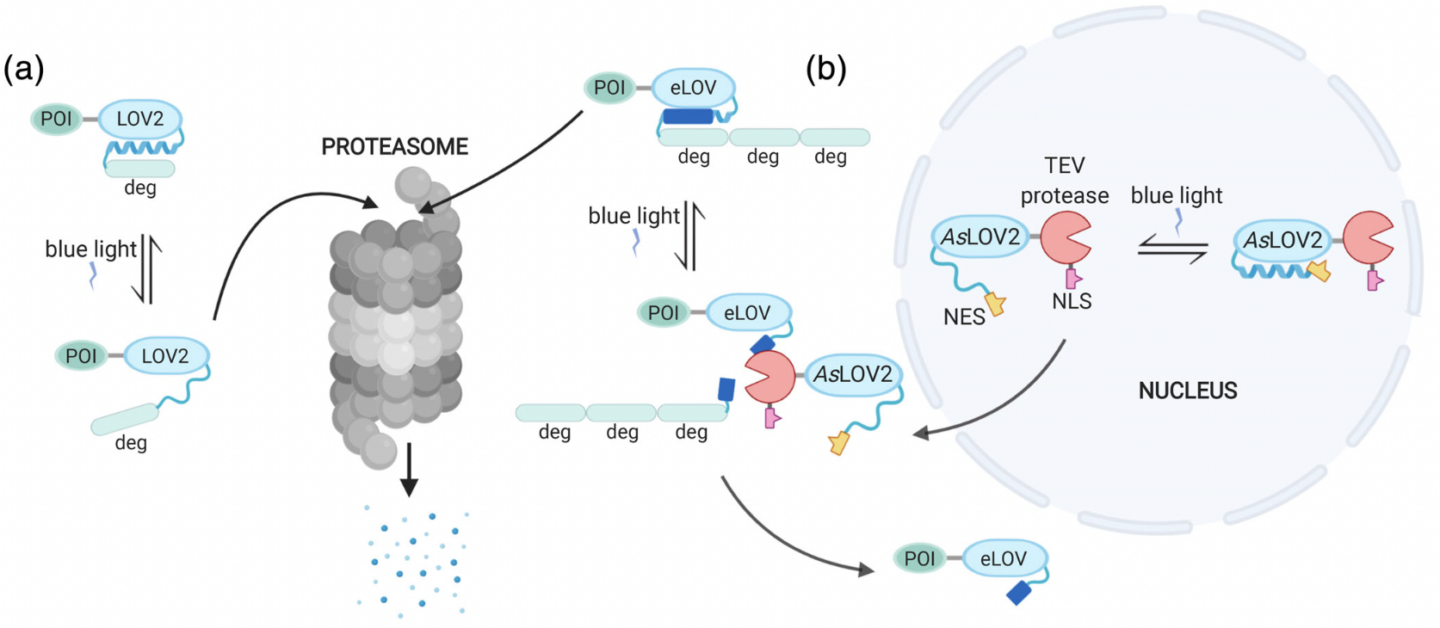
Kichuk, Therese C., César Carrasco‐López, and José L. Avalos. “Lights up on organelles: Optogenetic tools to control subcellular structure and organization”. WIREs Systems Biology and Medicine (2021). Web.
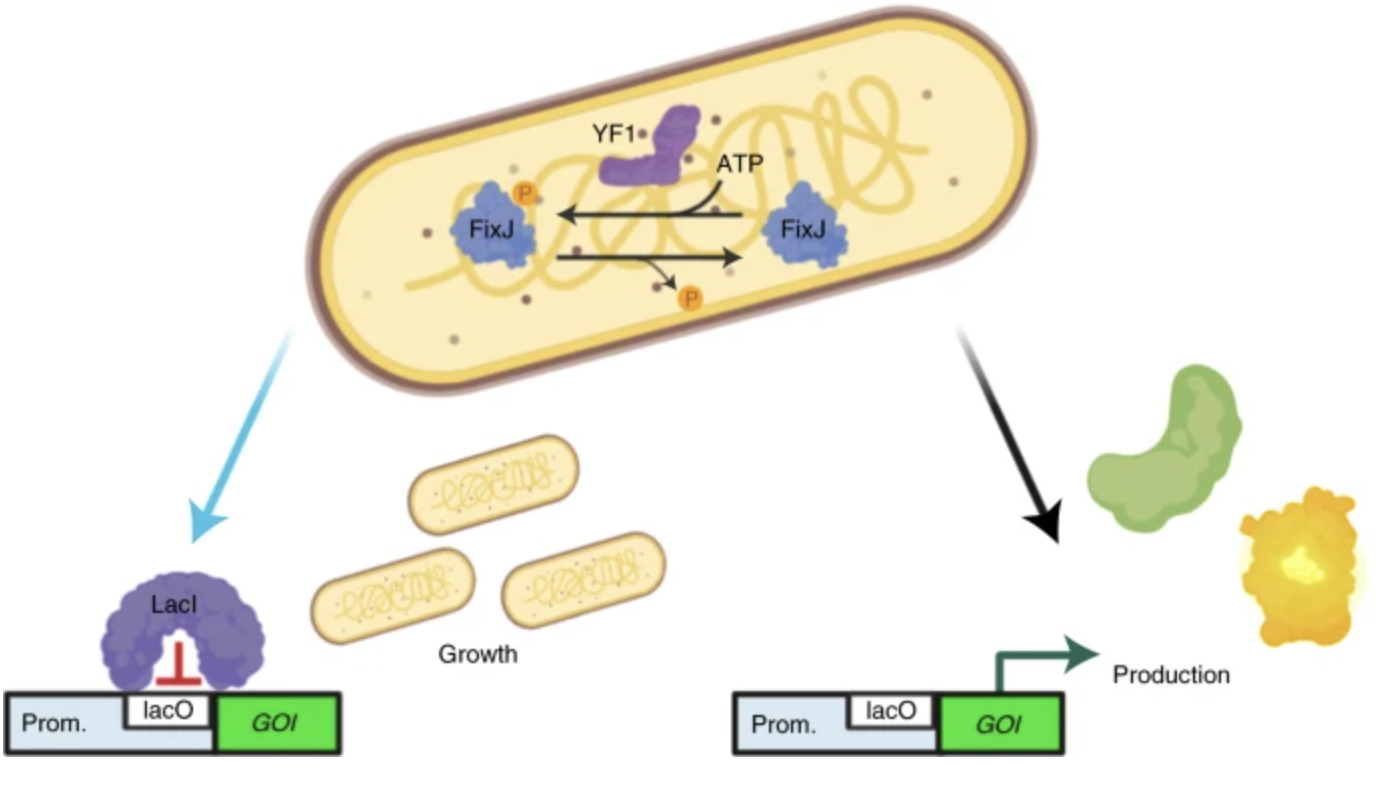
Lalwani, Makoto A., et al. “Optogenetic control of the lac operon for bacterial chemical and protein production”. Nature Chemical Biology (2021). Web.
2020
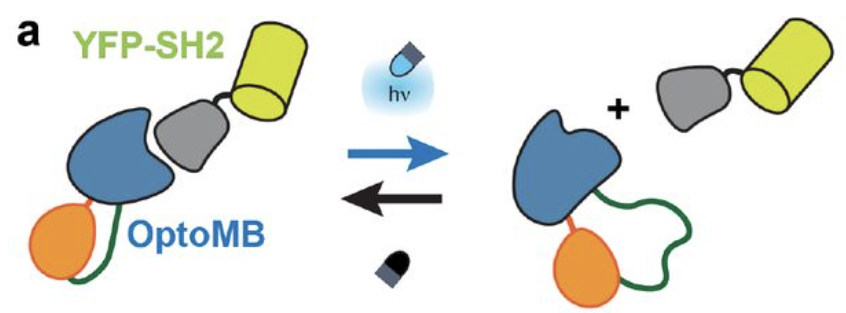
Carrasco-López, César, et al. “Light-responsive monobodies for dynamic control of customizable protein binding”. bioRxiv (2020): n. pag. Web.
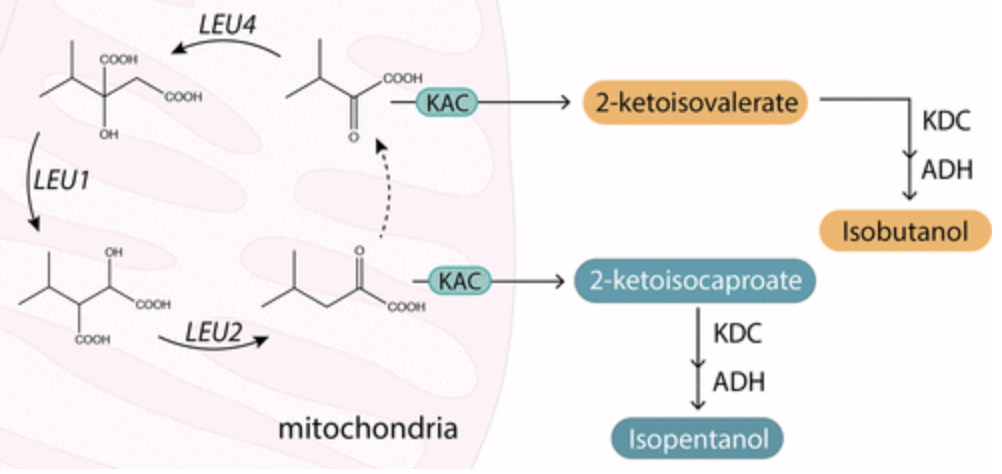
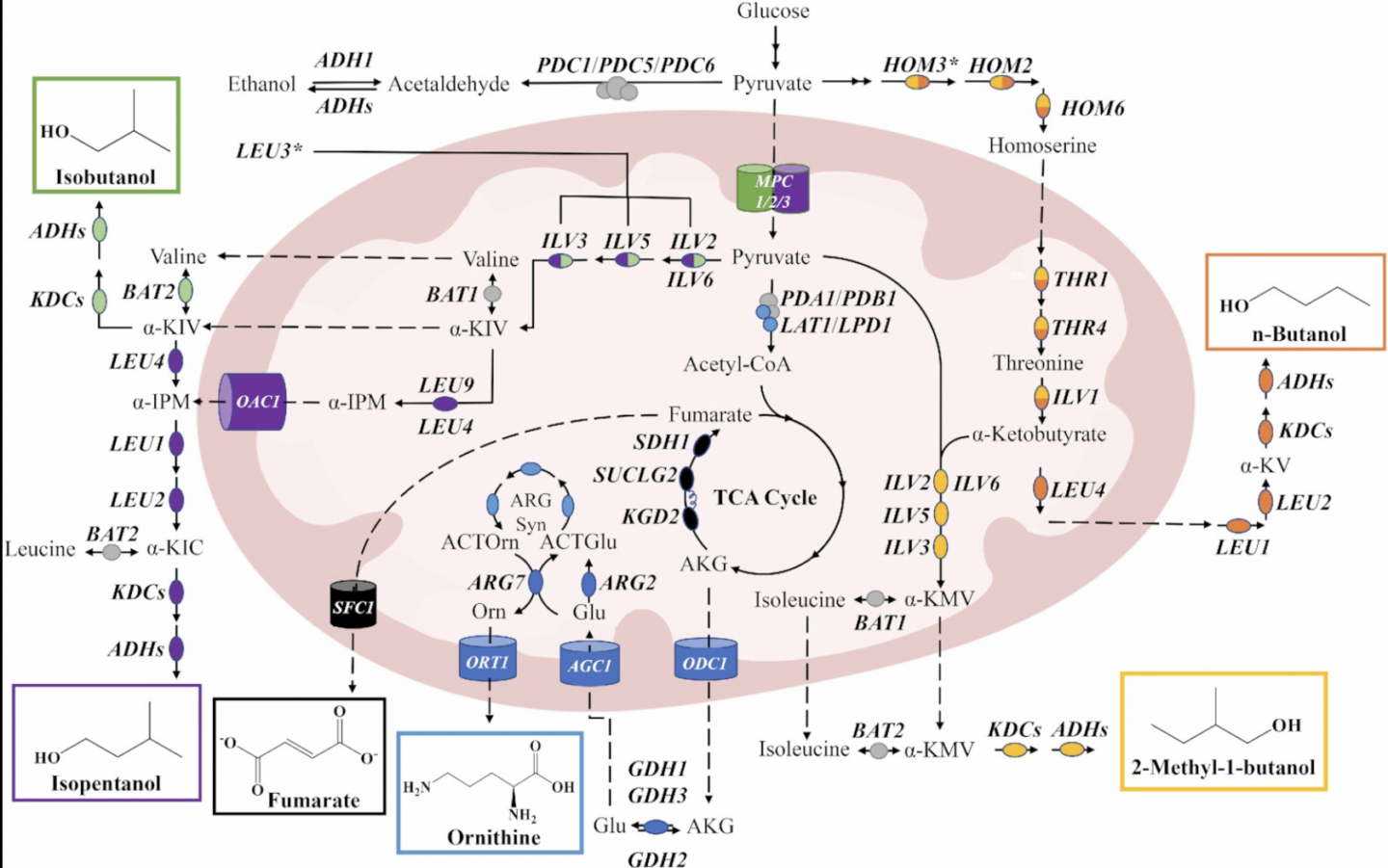
Hammer, Sarah K., Yanfei Zhang, and José L. Avalos. “Mitochondrial Compartmentalization Confers Specificity to the 2-Ketoacid Recursive Pathway: Increasing Isopentanol Production in Saccharomyces cerevisiae”. ACS Synthetic Biology (2020): n. pag. Web.
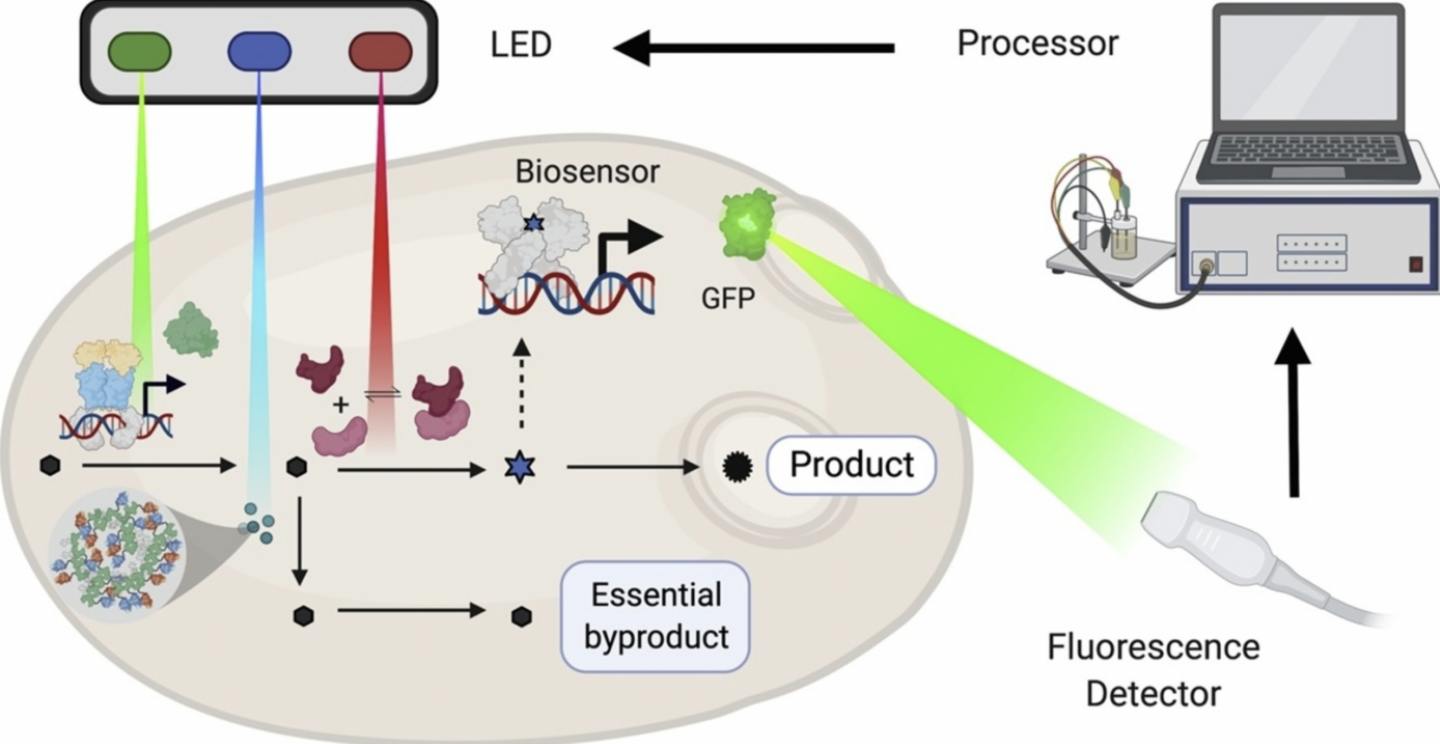
Carrasco-López, César, et al. “Optogenetics and biosensors set the stage for metabolic cybergenetics”. Current Opinion in Biotechnology 65 (2020). Web.
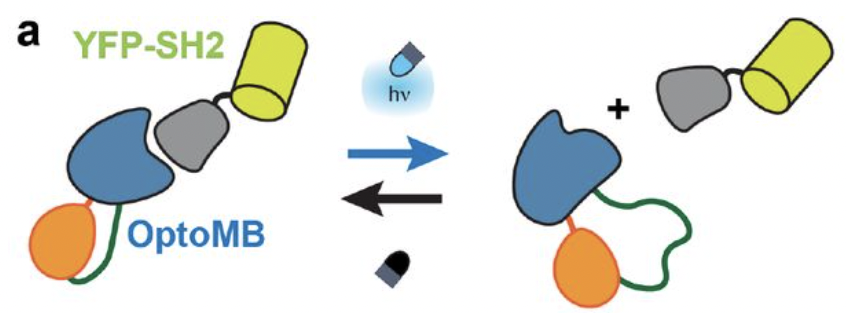
Carrasco-López, César, et al. “Development of light-responsive protein binding in the monobody non-immunoglobulin scaffold”. Nature Communications 11.1 (2020): , 11, 1, 1-13. Web.
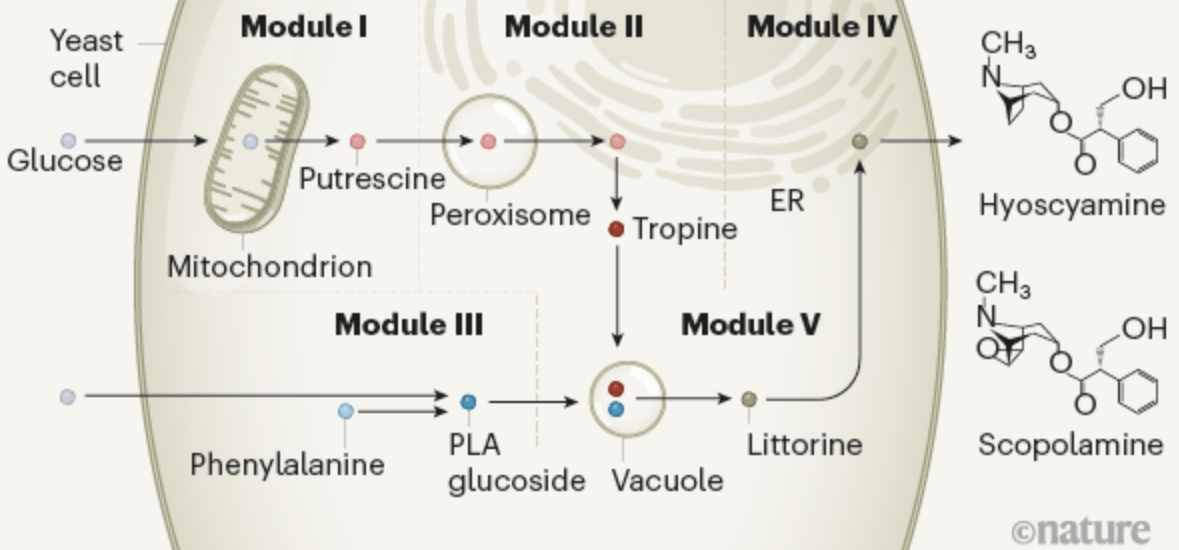
López, José Montaño, and José L. Avalos. “Genetically engineered yeast makes medicinal plant products”. Nature (2020). Web.
Duran, Lisset, José Montaño López, and José L. Avalos. “¡Viva la Mitochondria!: Harnessing yeast mitochondria for chemical production”. FEMS Yeast Research 20.6 (2020). Web.

Zhang, Yanfei, et al. “Genetically encoded biosensors for branched-chain amino acid metabolism to monitor mitochondrial and cytosolic production of isobutanol and isopentanol in yeast”. bioRxiv (2020): n. pag. Web.
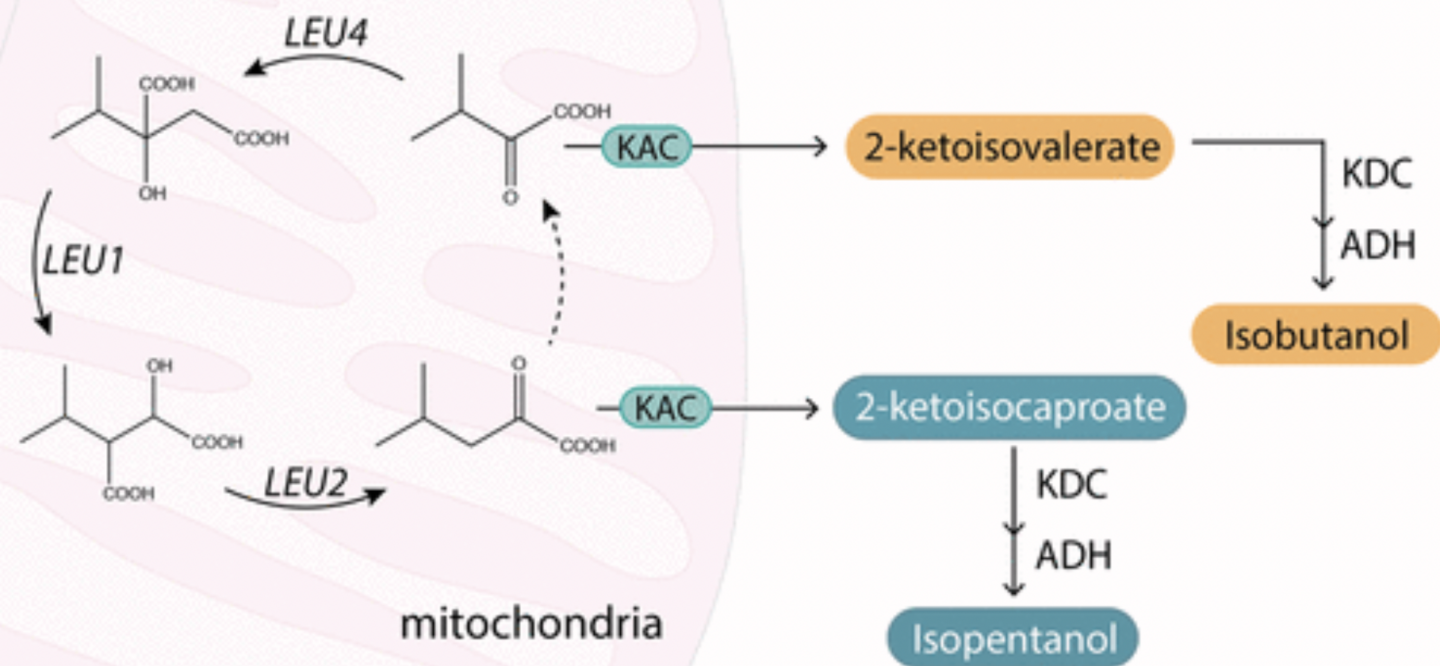
Hammer, Sarah K., Yanfei Zhang, and José L. Avalos. “Mitochondrial Compartmentalization Confers Specificity to the 2-Ketoacid Recursive Pathway: Increasing Isopentanol Production in Saccharomyces cerevisiae”. ACS Synthetic Biology (2020): n. pag. Web.
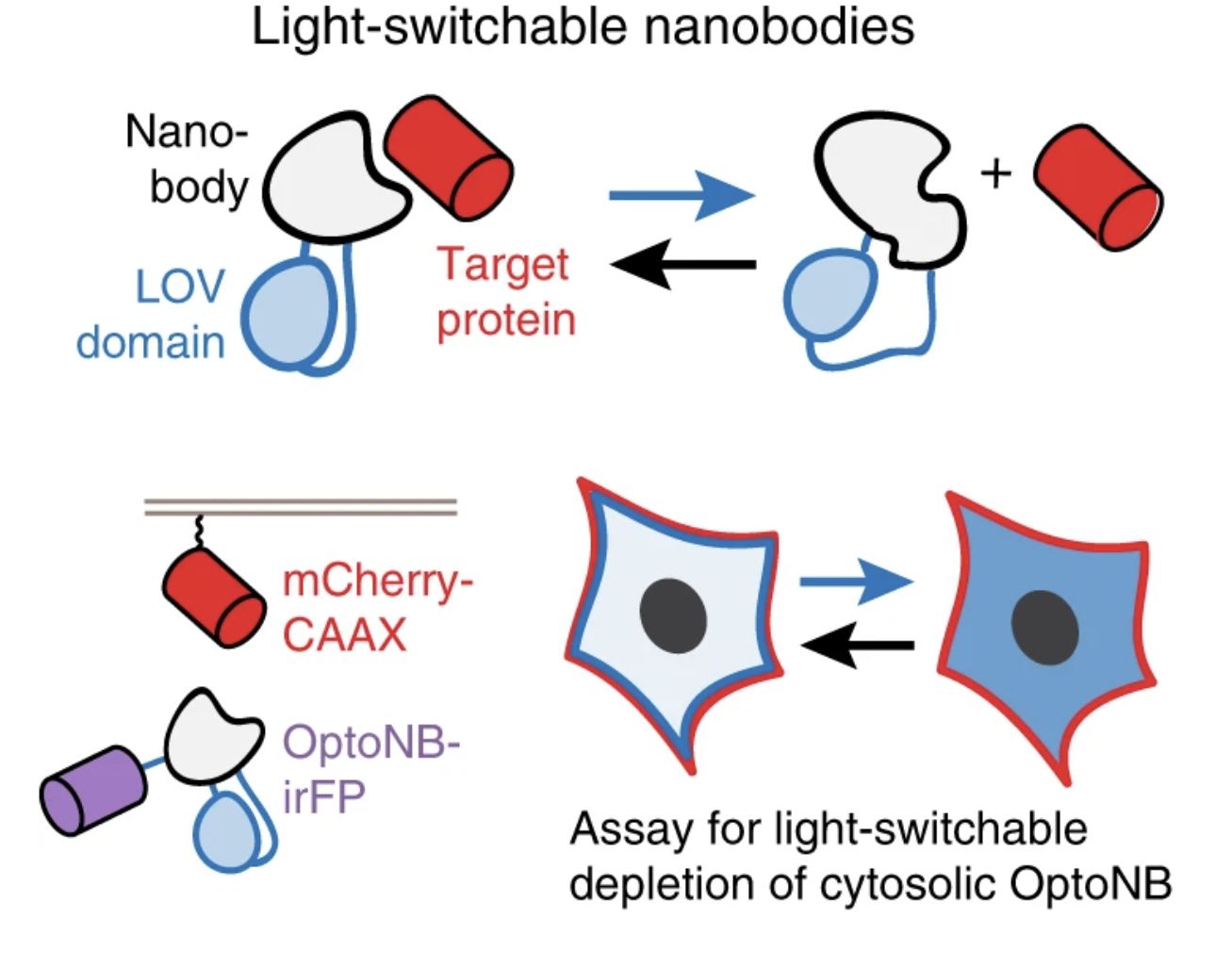
Gil, Agnieszka A., et al. “Optogenetic control of protein binding using light-switchable nanobodies”. Nature Communications 11.1 (2020): , 11, 1, 1-12. Web.
2019
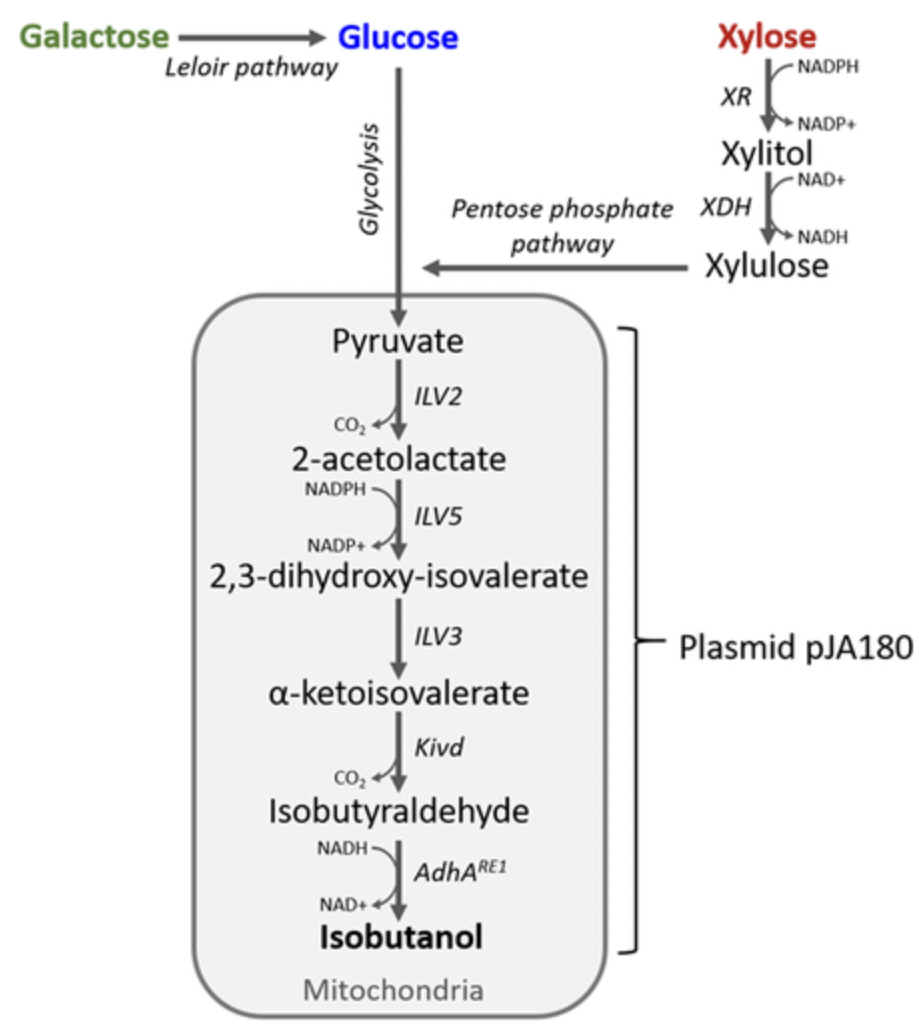
Lane, Stephan, et al. “Xylose assimilation enhances the production of isobutanol in engineered Saccharomyces cerevisiae”. Biotechnology and Bioengineering 2019. Web.
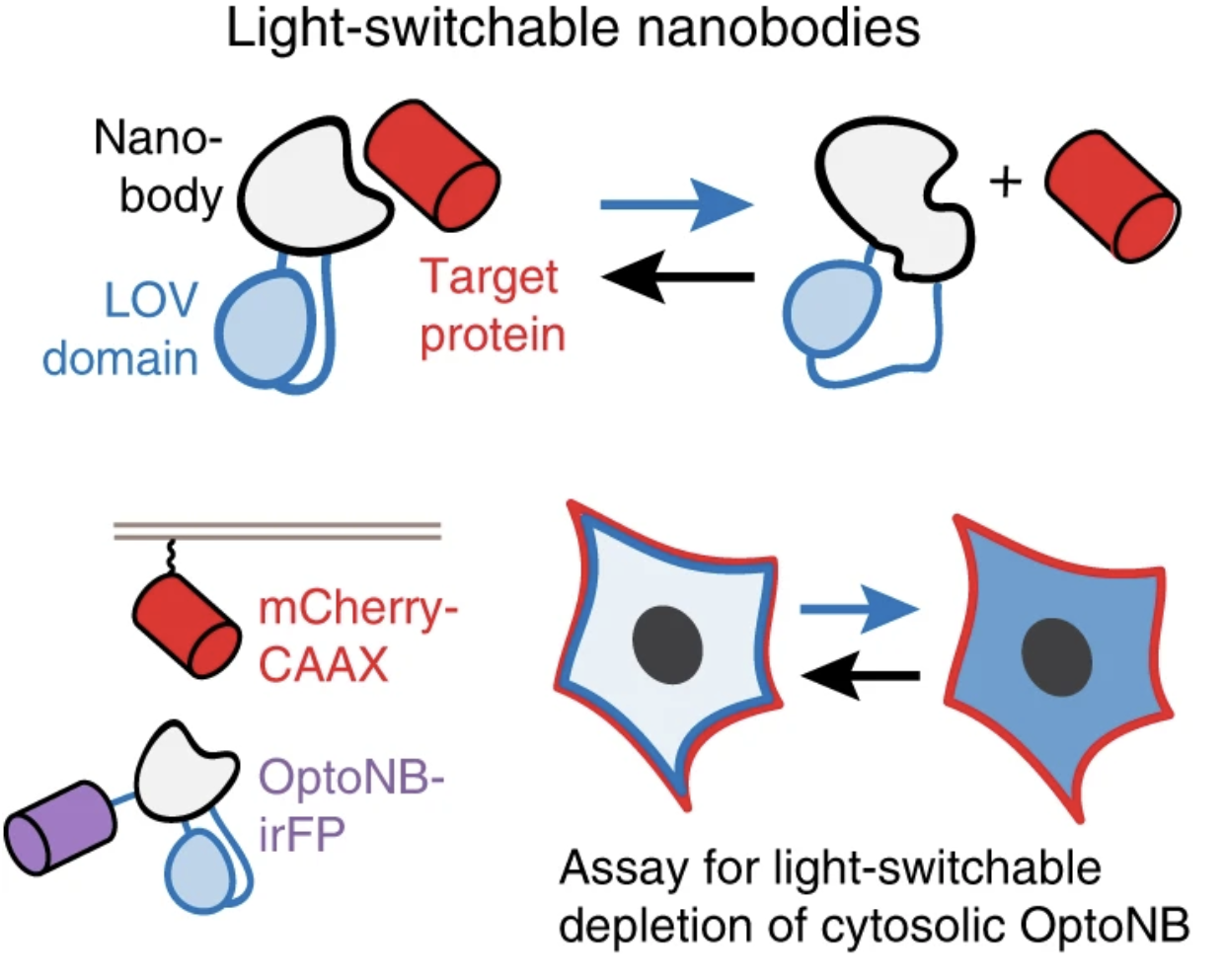
Gil, Agnieszka A., et al. “Optogenetic control of protein binding using light-switchable nanobodies”. bioRxiv 2019: n. pag. Print.
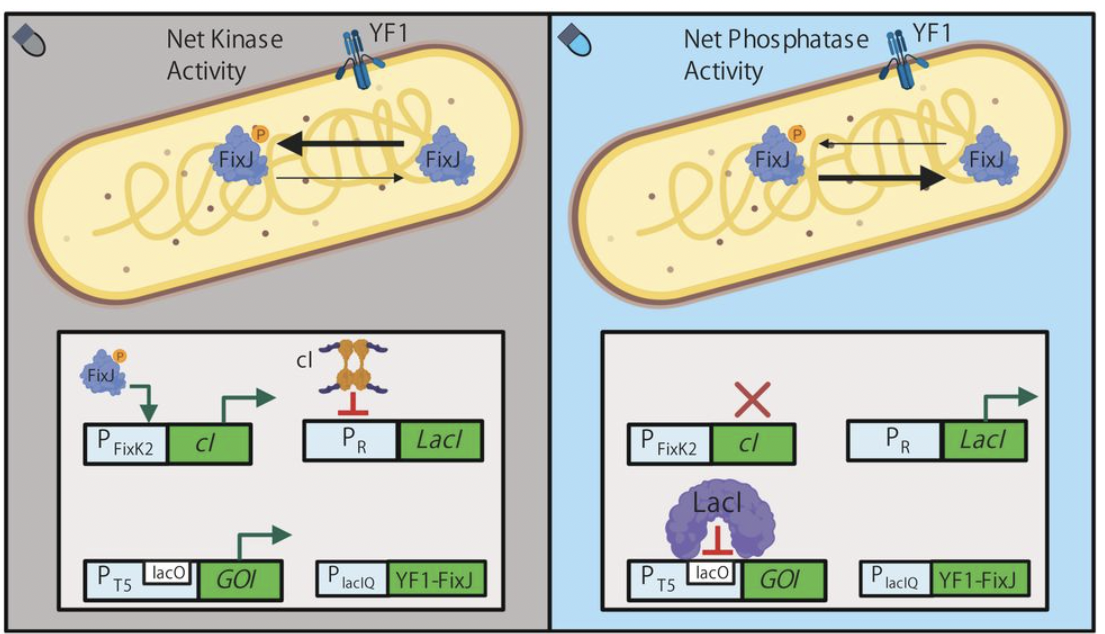
Lalwani, Makoto A., et al. “Optogenetic lac operon to control chemical and protein production in Escherichia coli with light”. bioRxiv 2019: n. pag. Print.
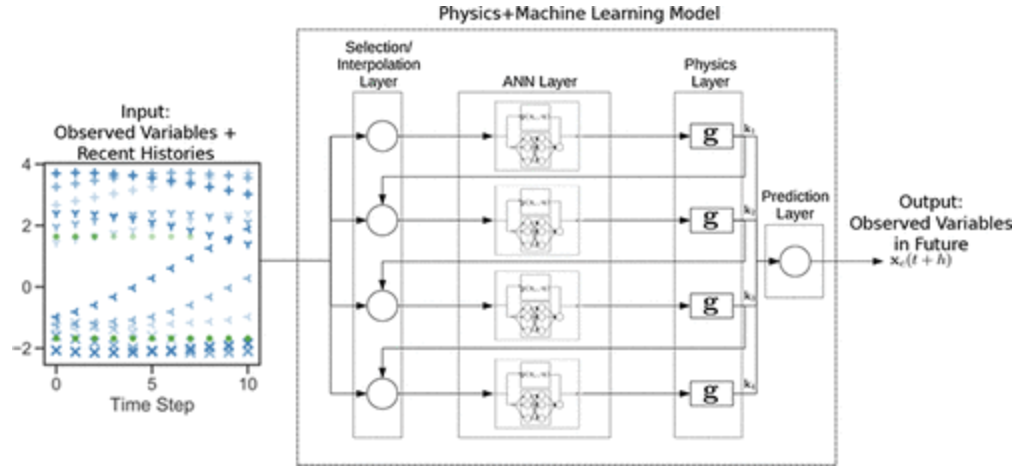
Lovelett, Robert, José L. Avalos, and Ioannis G. Kevrekidis. “Partial observations and conservation laws: Grey-box modeling in biotechnology and optogenetics”. Industrial & Engineering Chemistry Research (2019): n. pag. Print.
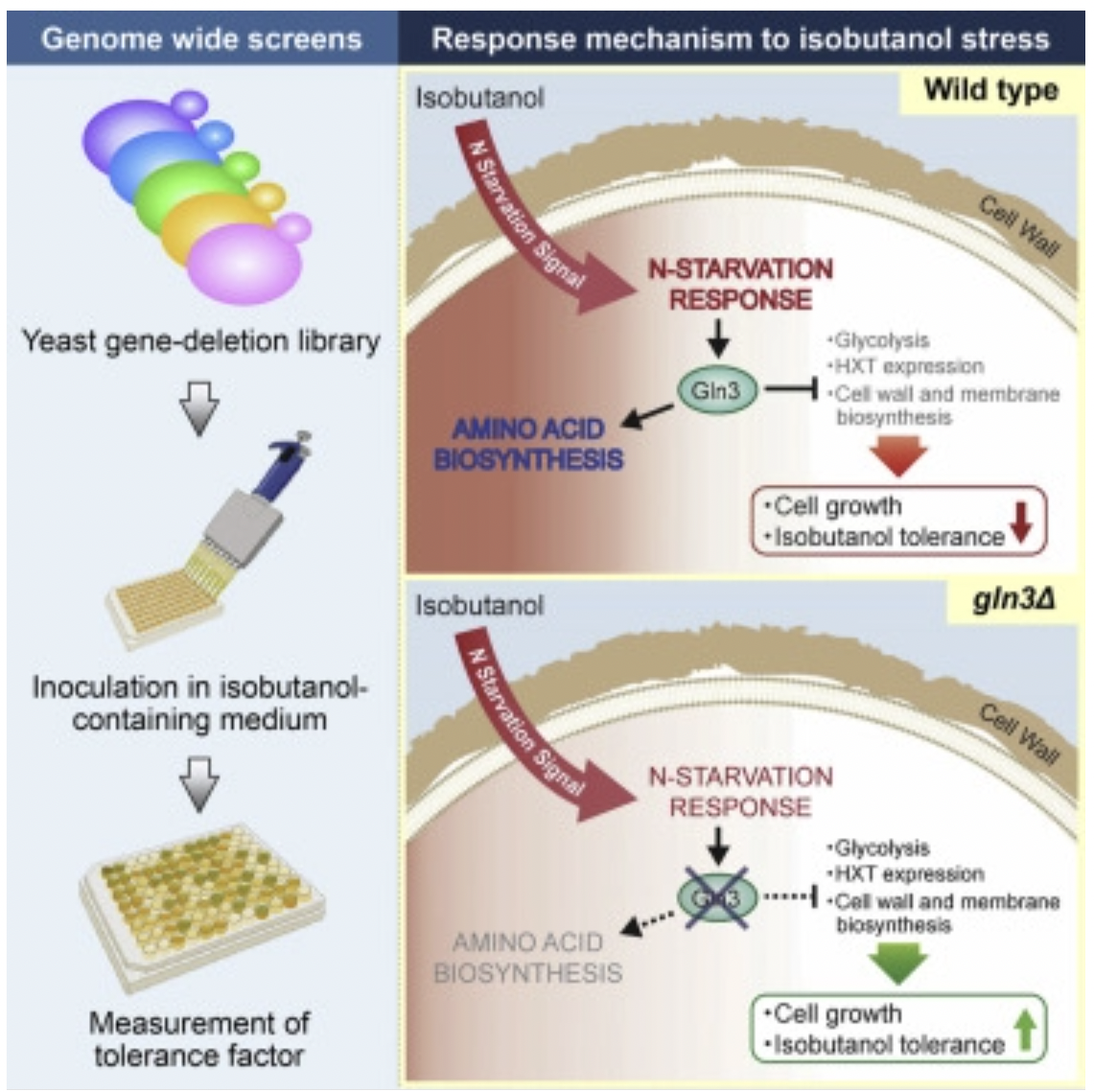
Kuroda, Kouichi, et al. “Critical Roles of the Pentose Phosphate Pathway and GLN3 in Isobutanol-Specific Tolerance in Yeast”. Cell Systems (2019): n. pag. Print.
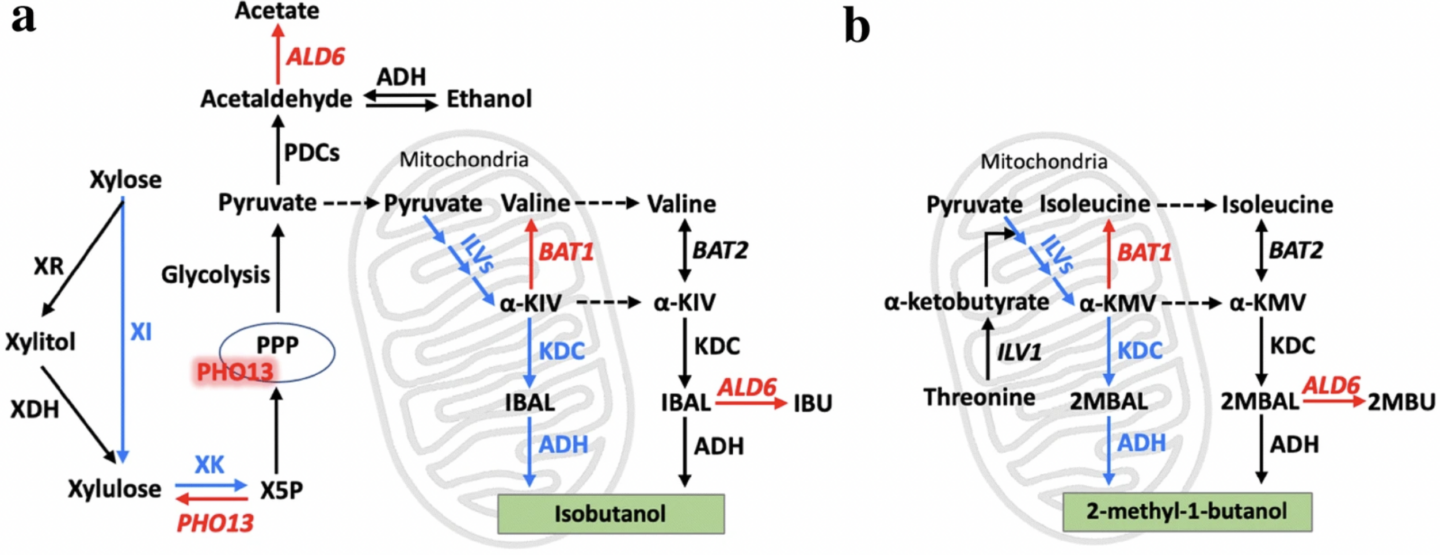
Zhang, Yanfei, et al. “Xylose utilization stimulates mitochondrial production of isobutanol and 2-methyl-1-butanol in Saccharomyces cerevisiae”. Biotechnology for Biofuels (2019): n. pag. Print.
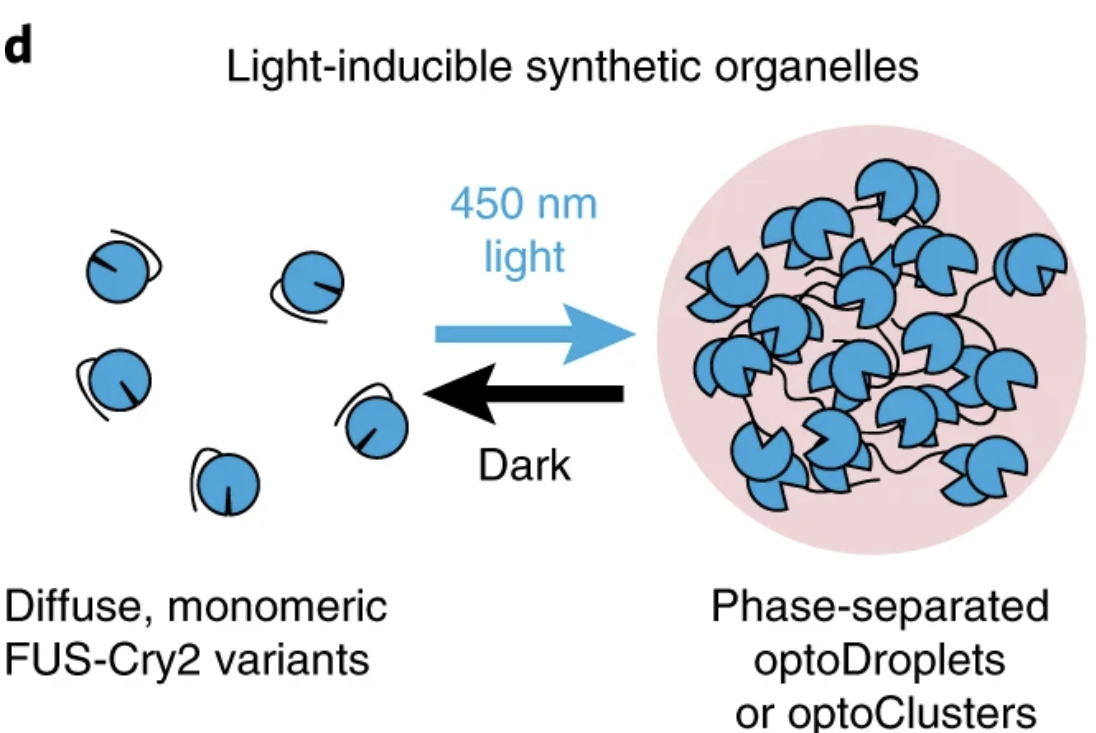
Zhao, Evan M., et al. “Light-based control of metabolic flux through assembly of synthetic organelles”. Nature Chemical Biology (2019). Web.
2018
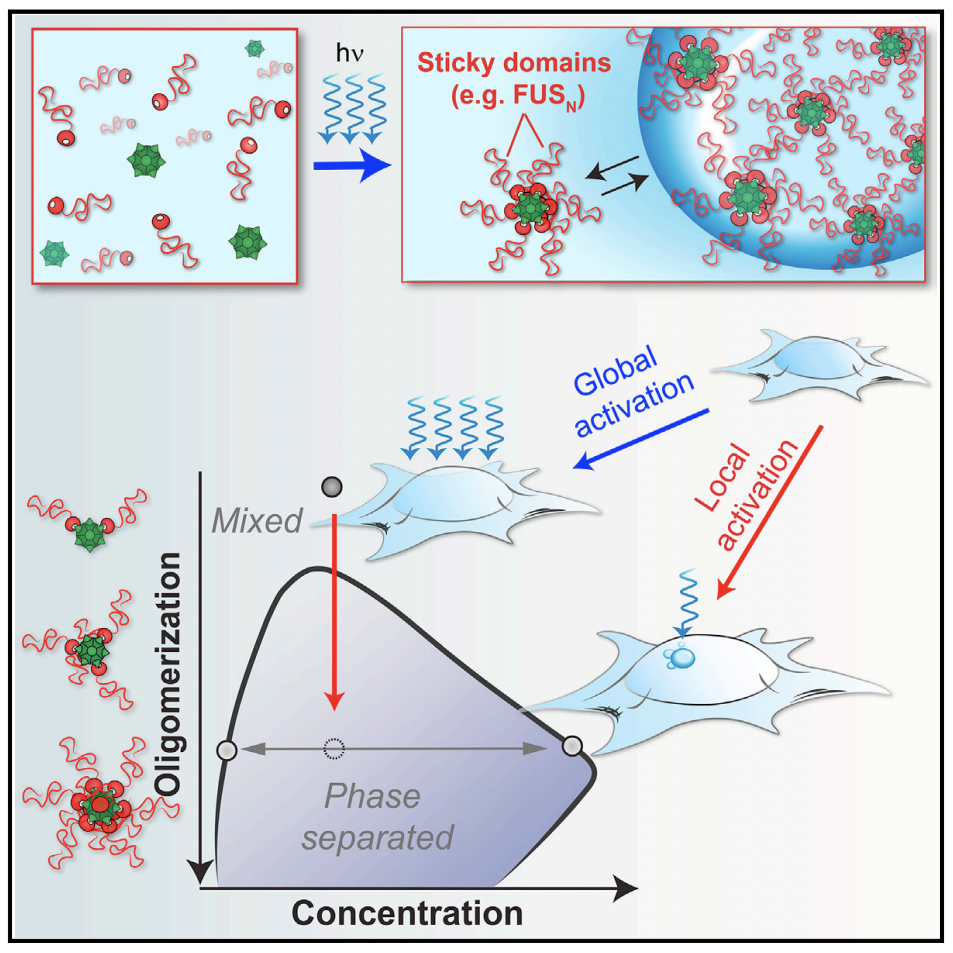
Bracha, Dan, et al. “Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds”. Cell 175.6 (2018): , 175, 6, 1467–1480. Web.
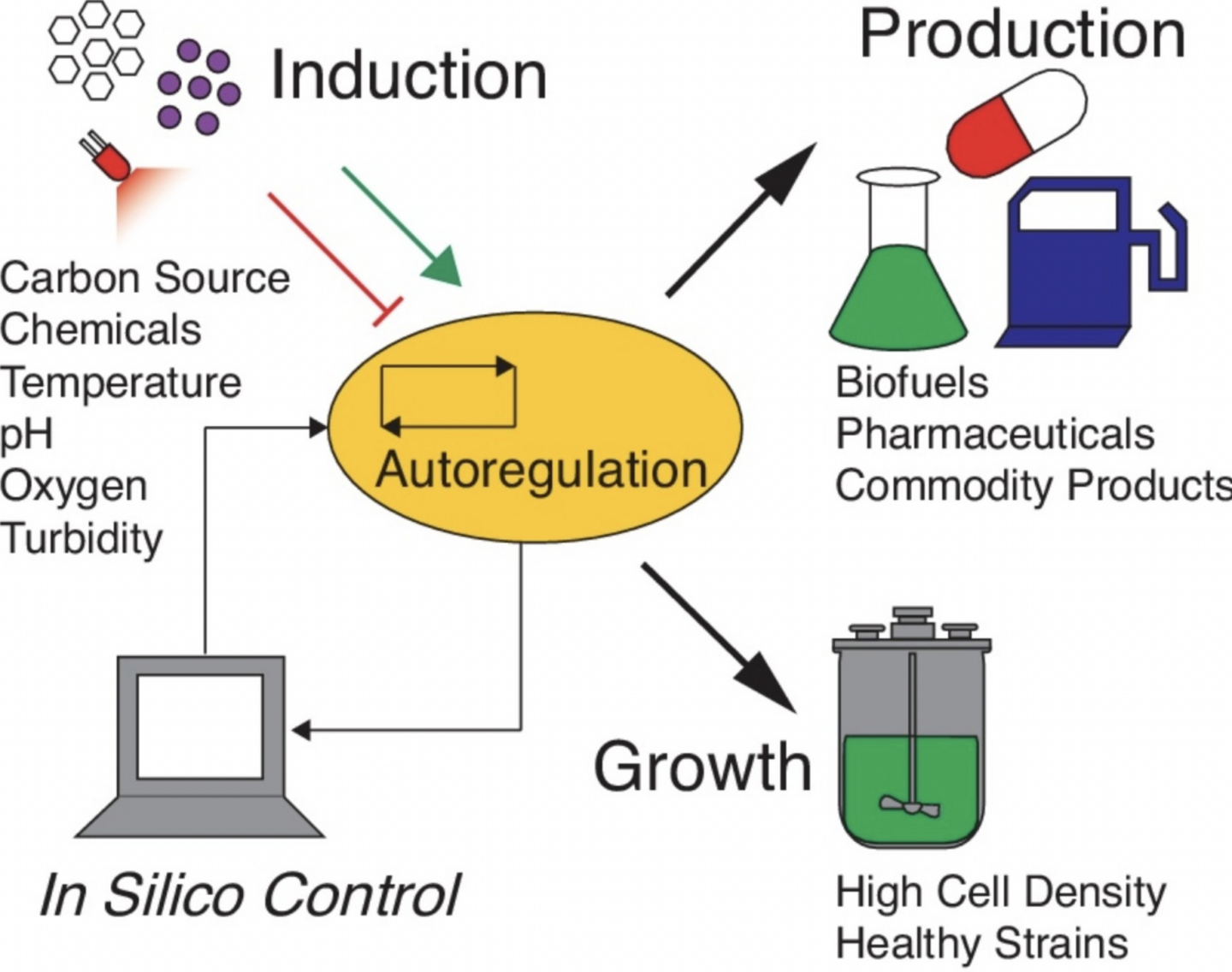
Lalwani, Makoto A., Evan M. Zhao, and José L. Avalos. “Current and future modalities of dynamic control in metabolic engineering”. Current Opinion in Biotechnology (2018). Web.
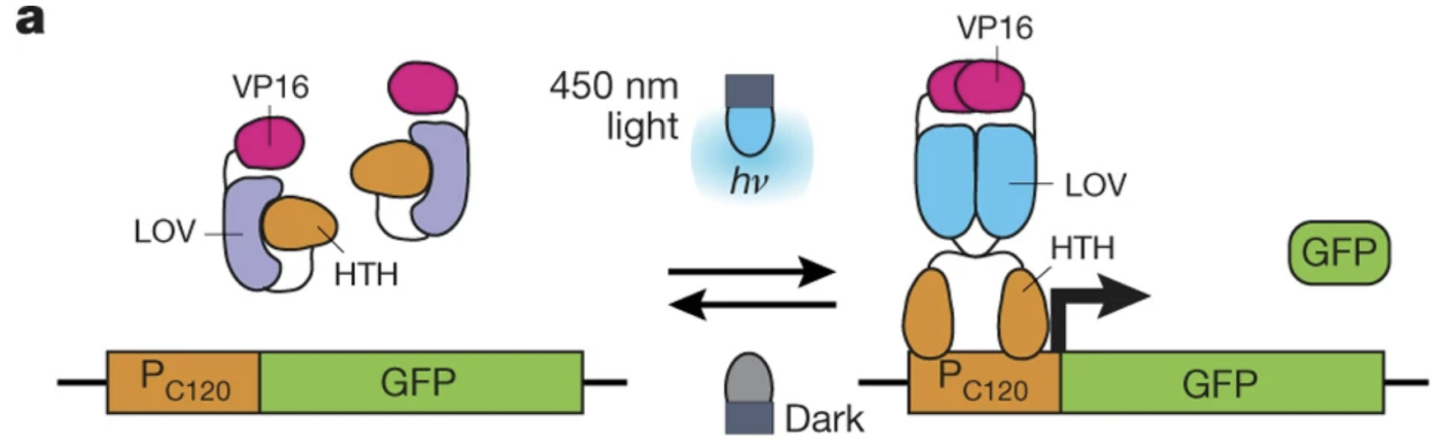
Zhao, Evan M., et al. “Optogenetic regulation of engineered cellular metabolism for microbial chemical production”. Nature (2018). Web.
2017
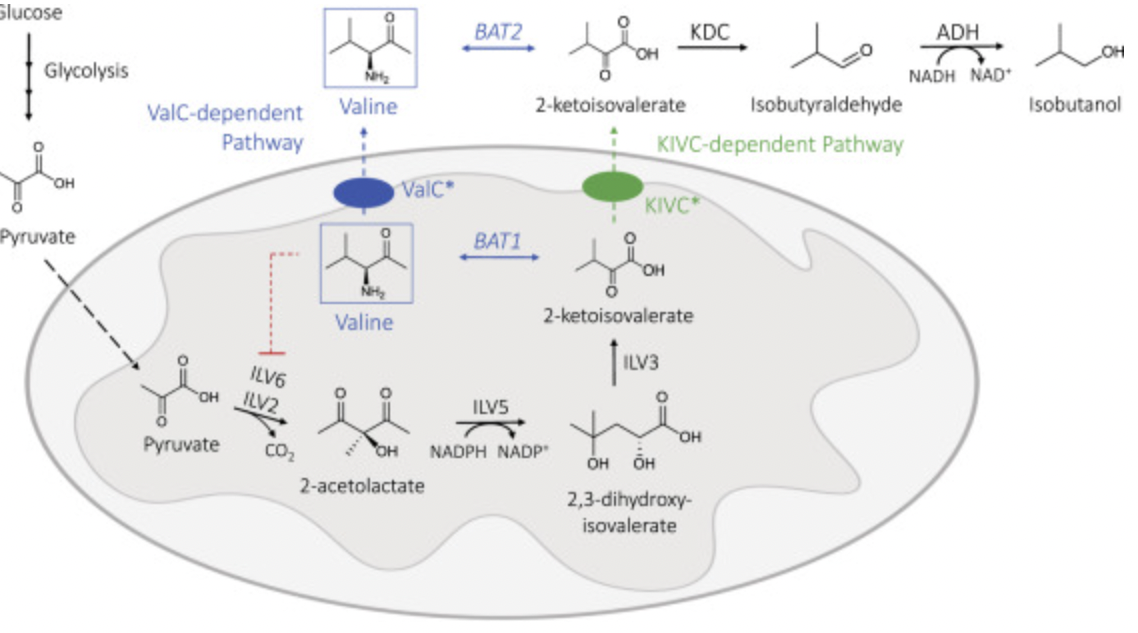
Hammer, Sarah K., and José L. Avalos. “Uncovering the role of branched-chain amino acid transaminases in Saccharomyces cerevisiae isobutanol biosynthesis”. Metabolic Engineering (2017): , -. Web.
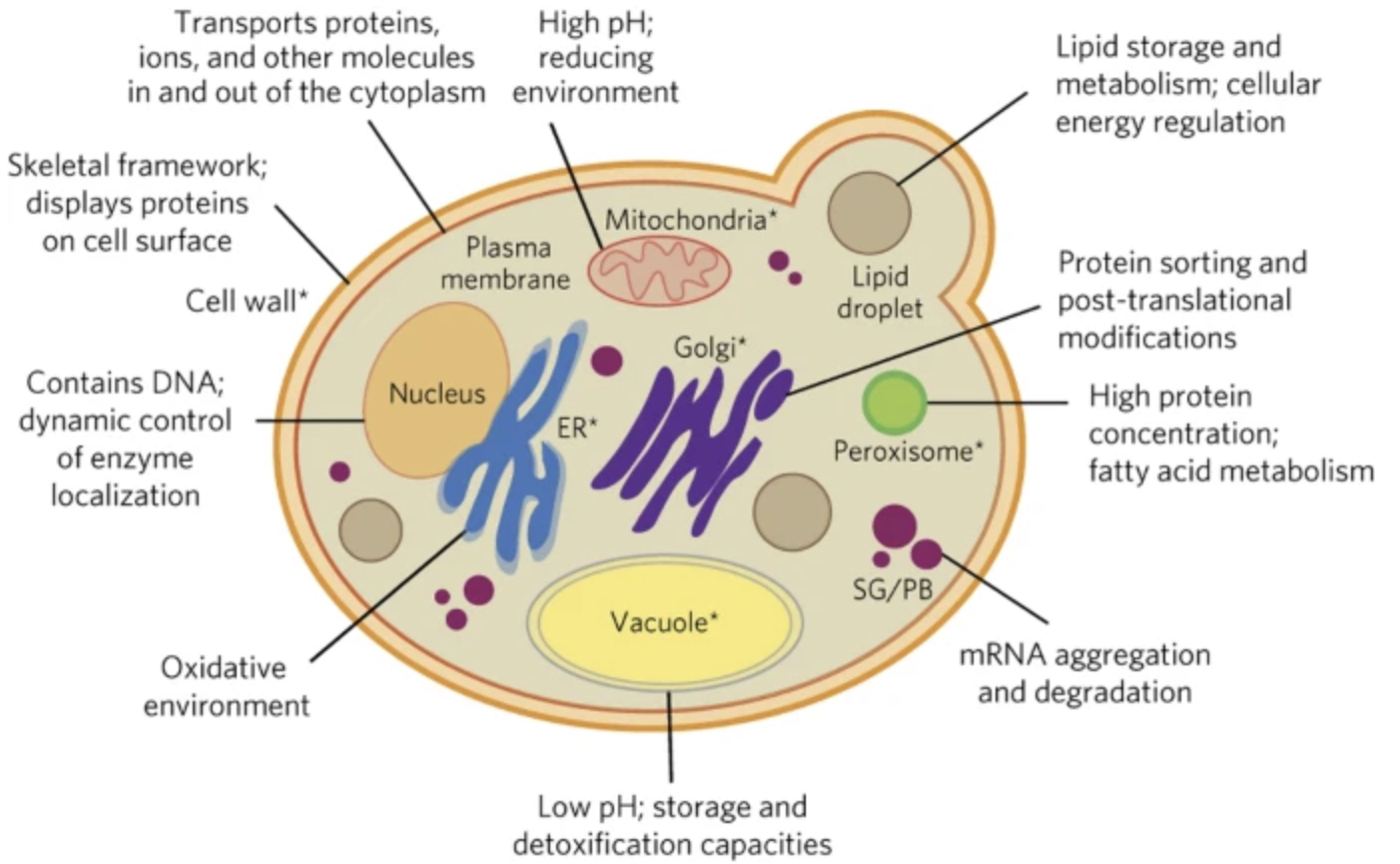
Hammer, Sarah K., and José L. Avalos. “Harnessing yeast organelles for metabolic engineering”. Nature Chemical Biology (2017): , 823–832. Web.
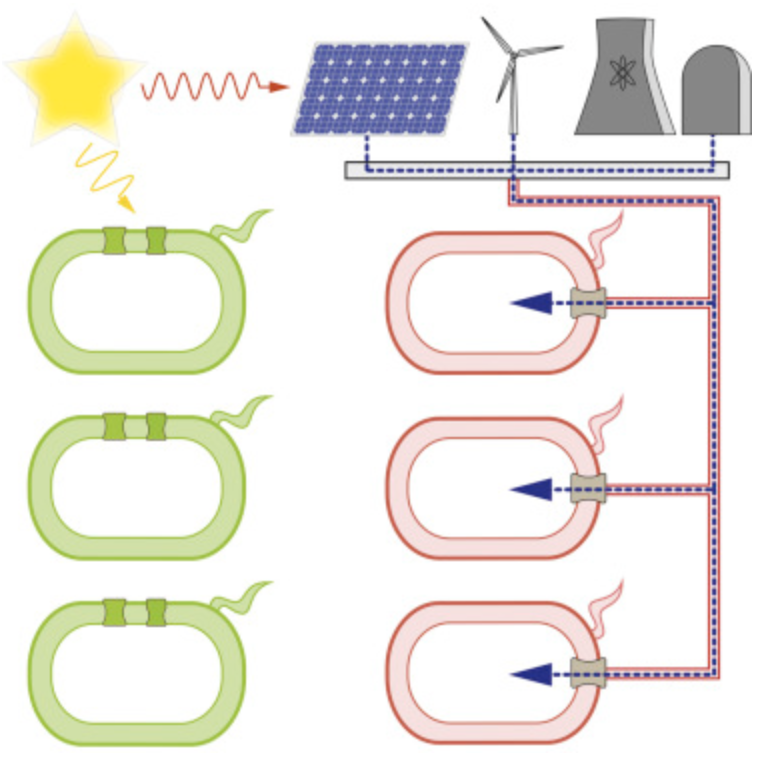
Adesina, Oluwakemi, et al. “Embracing Biological Solutions to the Sustainable Energy Challenge”. Chem 21 (2017): , 2, 1, 20 – 51. Web.
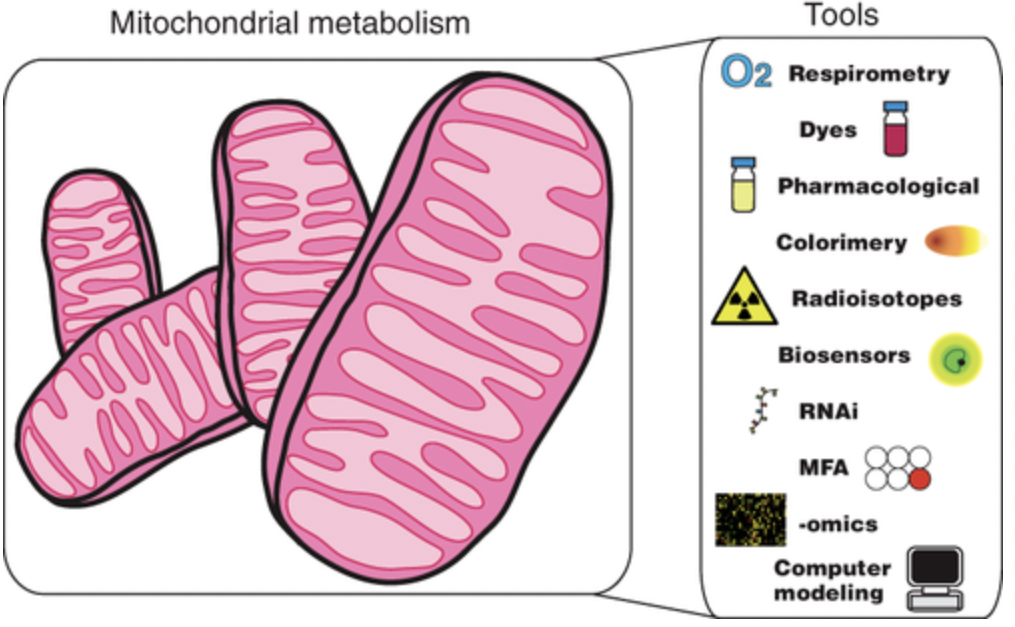
Zhang, Yanfei, and José L. Avalos. “Traditional and novel tools to probe the mitochondrial metabolism in health and disease”. Wiley Interdisciplinary Reviews: Systems Biology and Medicine 92 (2017). Web.
2016
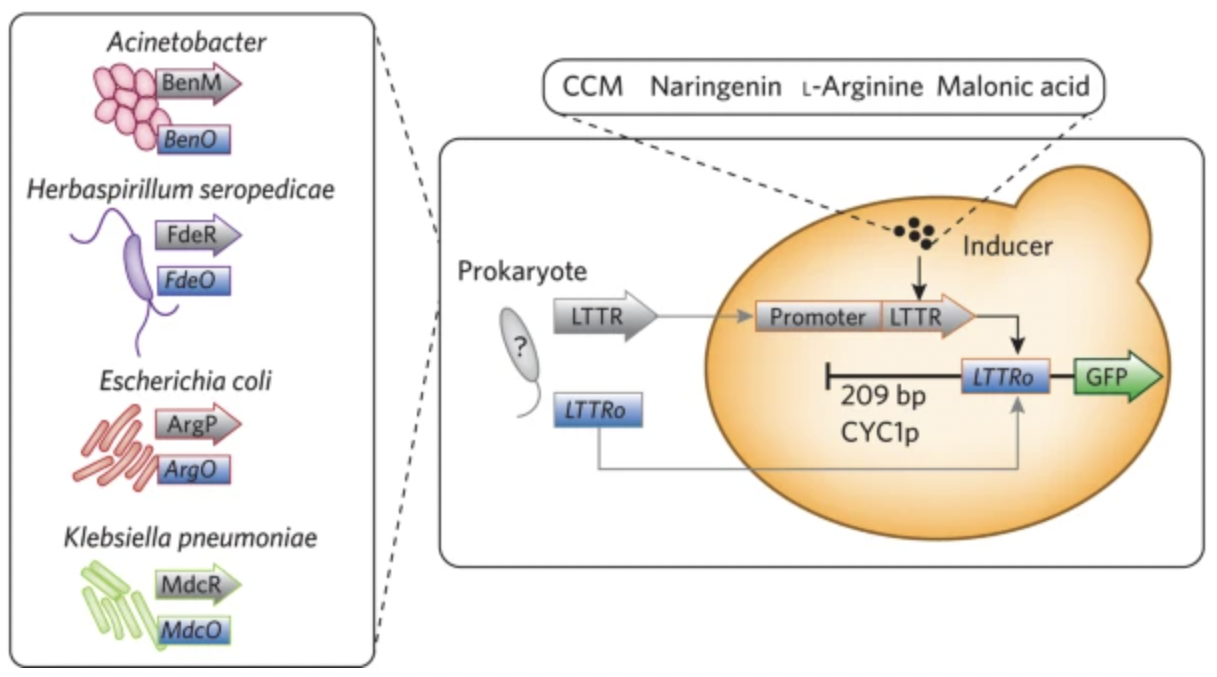
Hammer, Sarah K., and Jose L. Avalos. “Metabolic engineering: Biosensors get the green light”. Nat Chem Biol 2016894-895. Web.
2013
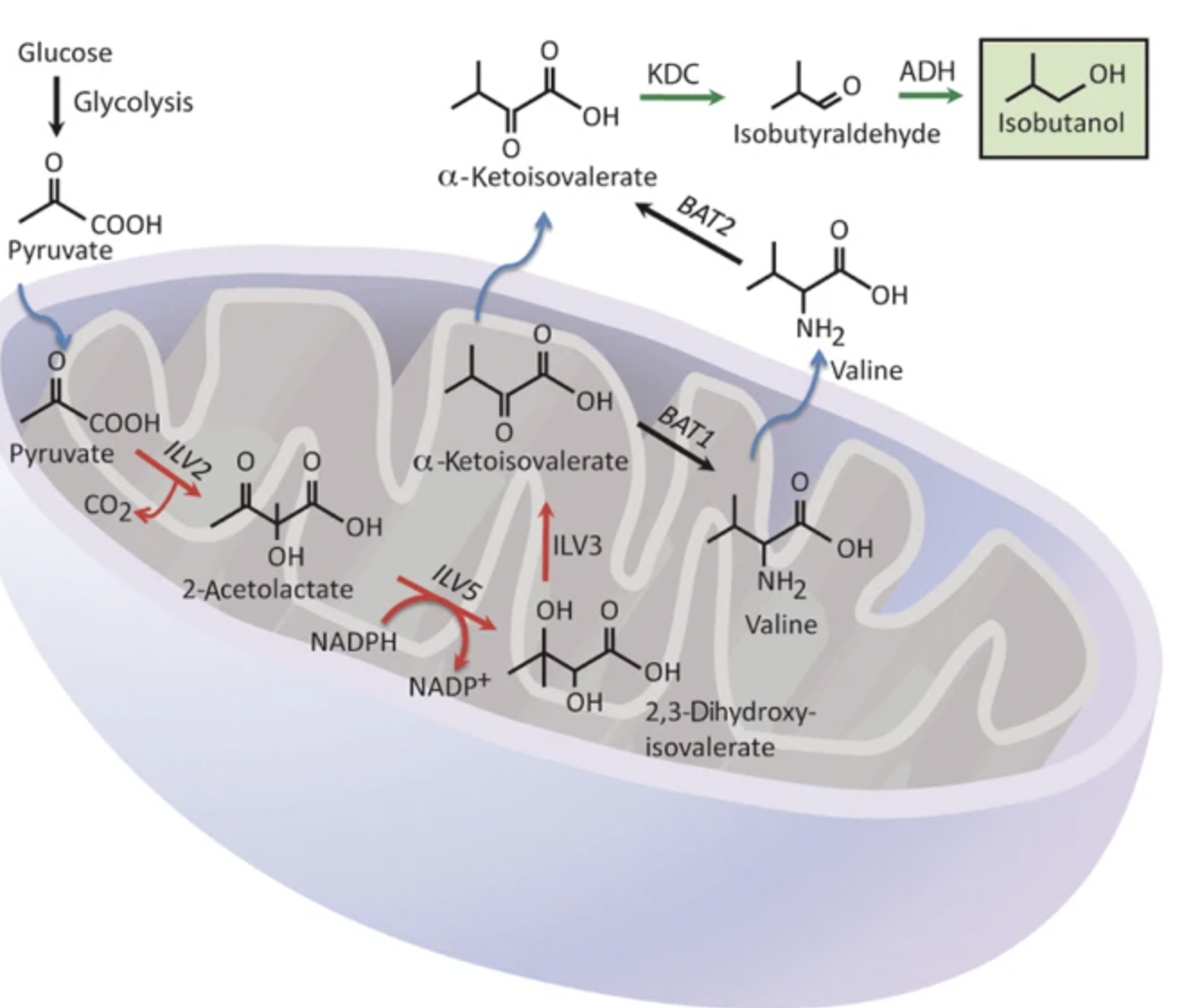
Avalos, Jose L., Gerald R. Fink, and Gregory Stephanopoulos. “Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols”. Nature Biotechnology 31 (2013): , 31, 335-341. Web.
2009
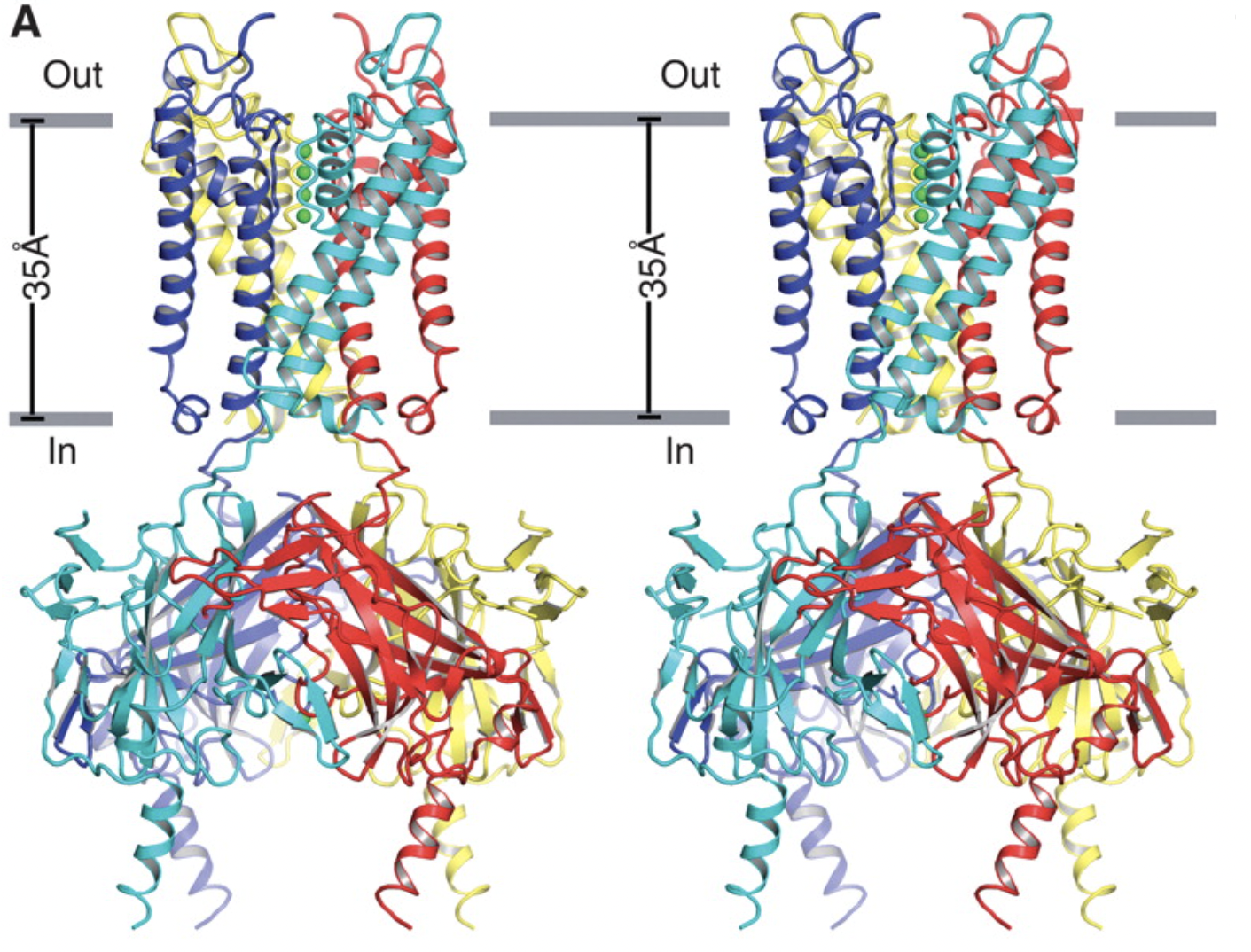
Tao, Xiao, et al. “Crystal Structure of the Eukaryotic Strong Inward-Rectifier K+ Channel Kir2.2 at 3.1 angstrom Resolution”. Science 326 (2009): , 326, 1668-1674. Web.
2006
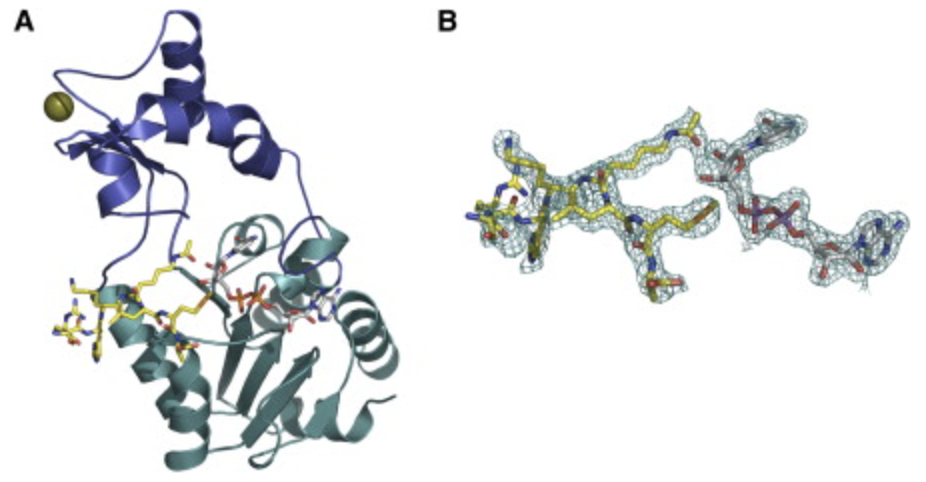
Hoff, Kevin G., et al. “Insights into the sirtuin mechanism from ternary complexes containing NAD(+) and acetylated peptide”. Structure 14 (2006): , 14, 1231-1240. Web.
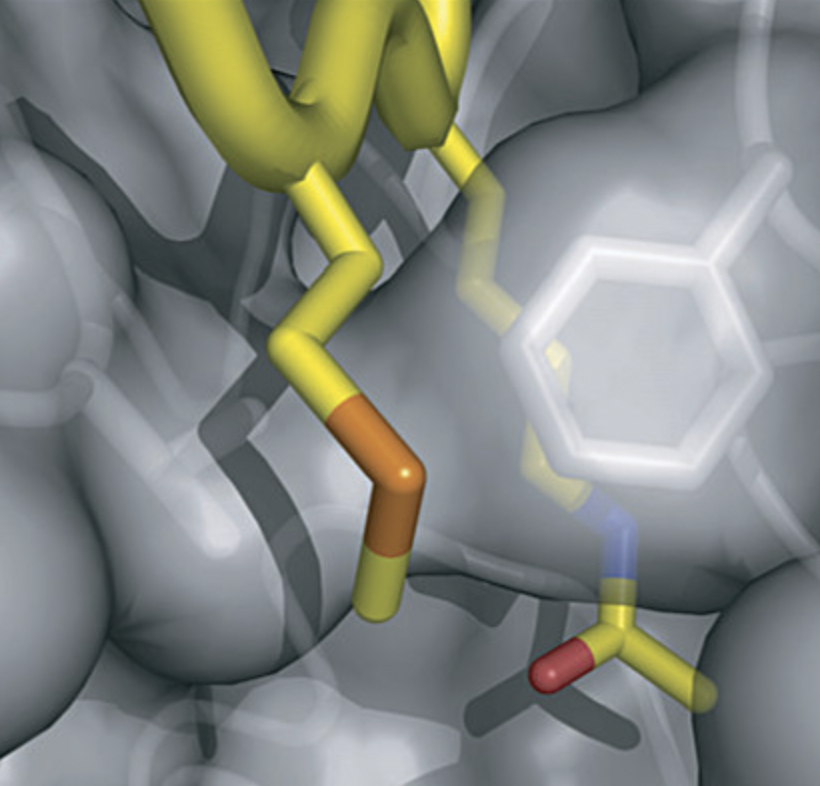
Cosgrove, M. S., et al. “The structural basis of sirtuin substrate affinity”. Biochemistry 45 (2006): , 45, 7511-7521. Web.
2005
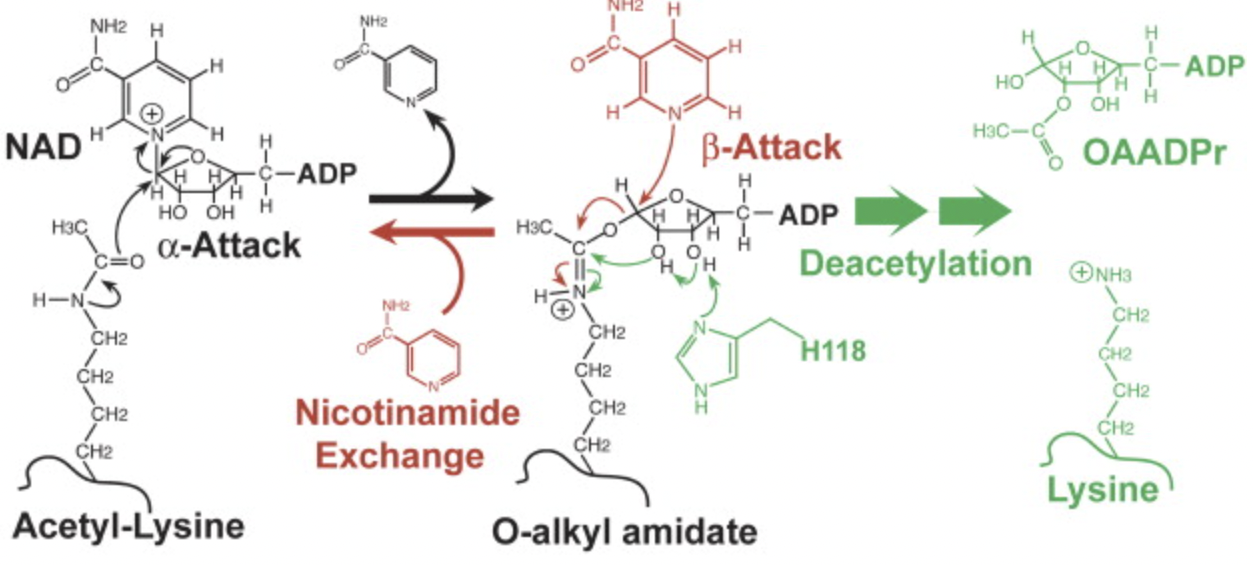
Avalos, J. L., K. M. Bever, and C. Wolberger. “Mechanism of sirtuin inhibition by nicotinamide: Altering the NAD(+) cosubstrate specificity of a Sir2 enzyme”. Molecular Cell 17 (2005): , 17, 855-868. Web.
2004
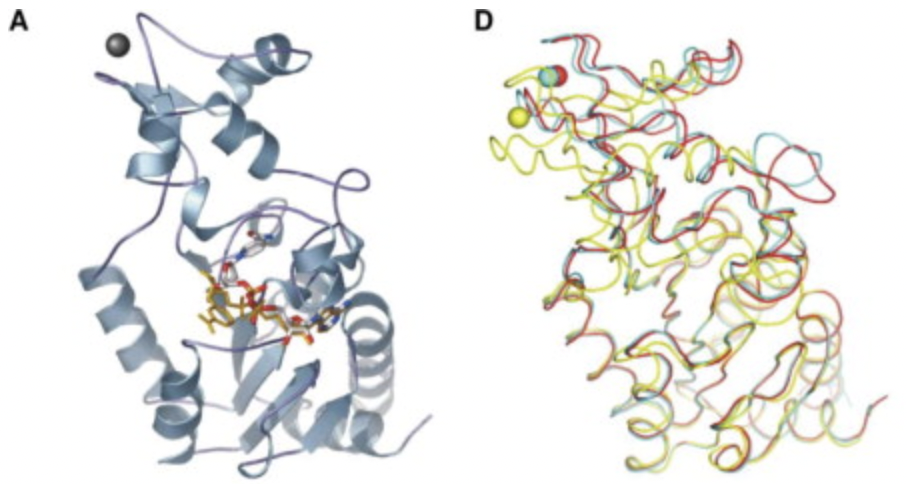
Avalos, J. L., J. D. Boeke, and C. Wolberger. “Structural basis for the mechanism and regulation of Sir2 enzymes”. Molecular Cell 13 (2004): , 13, 639-648. Web.
2002
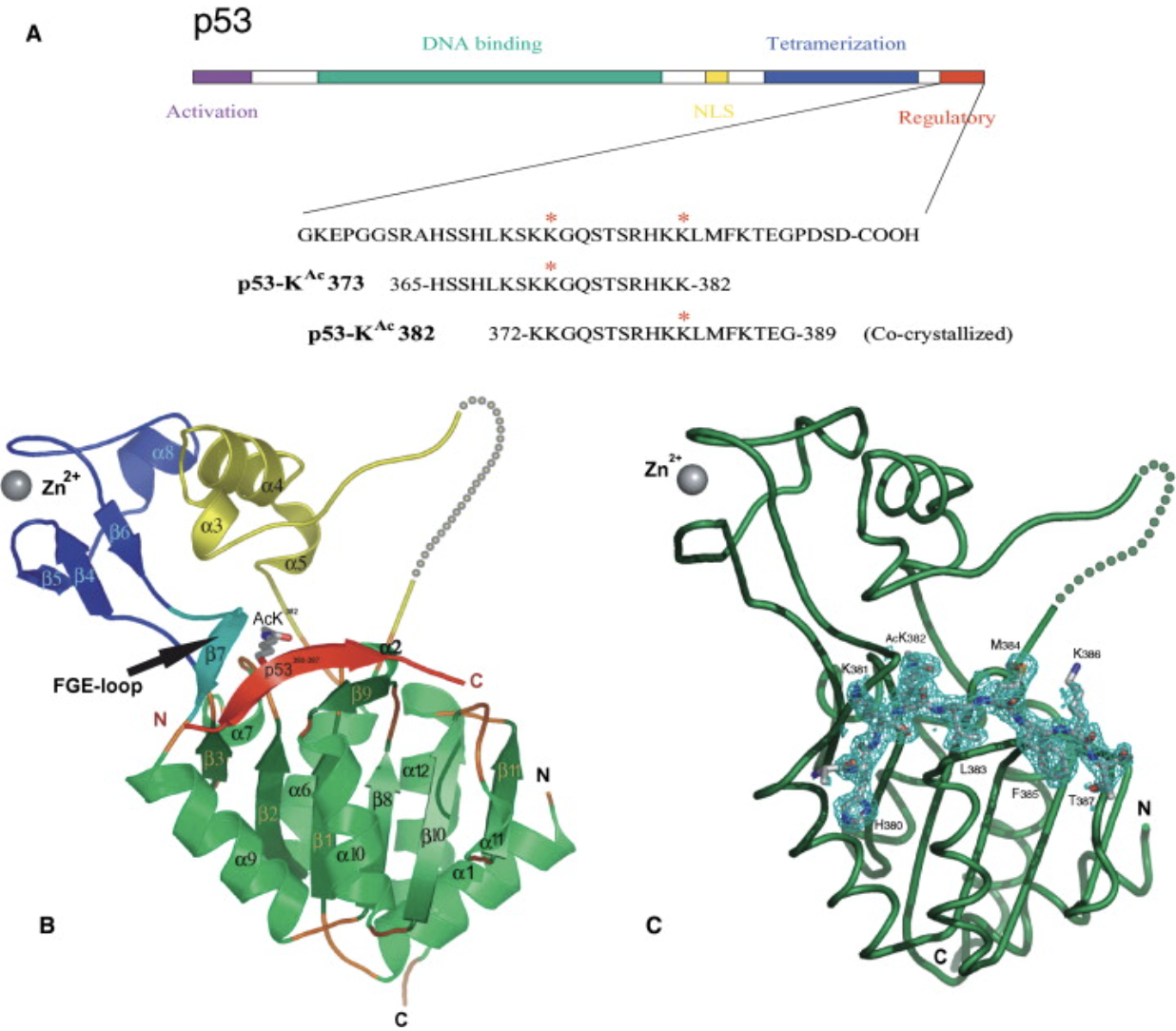
Avalos, J. L., et al. “Structure of a Sir2 enzyme bound to an acetylated p53 peptide”. Molecular Cell 10 (2002): , 10, 523-535. Web.
2000
Smith, J. S., et al. “A phylogenetically conserved NAD(+)-dependent protein deacetylase activity in the Sir2 protein family”. Proceedings of the National Academy of Sciences of the United States of America 97 (2000): , 97, 6658-6663. Web.